, Sandra van’t Padje1, John C. van Swieten1 and Ben A. Oostra1
(1)
Departments of Clinical Genetics and Neurology, Erasmus MC, Rotterdam, The Netherlands
Abstract
Zebrafish, a freshwater tropical fish, is a premiere model organism to study vertebrate development. Fast external development and transparency during embryogenesis allow for visual screening at the macroscopical and microscopical level, including visualization of organogenesis. High fecundity and short generation times facilitate genetic analyses. Zebrafish may be a particular powerful model for the study of human disease because many cellular processes are conserved throughout vertebrate evolution, including the corresponding disease genes. Finally, the ability to manipulate gene expression has broad usefulness in the study of modeling human disease, including dementia.
Key words
ZebrafishMorpholinoknock downtransgenicTILLINGdementia1 Introduction
The Latin name for zebrafish is Danio (formerly Brachydanio) rerio, which originates from the river Ganges in India and is a common aquarium fish throughout the world (1). Zebrafish belong to the cyprinid family of teleost fish (Fig. 1). In 1981, George Streisinger introduced the freshwater tropical zebrafish as a genetic model to study vertebrate development. Because of its transparent embryo that develops outside the mother’s body, the zebrafish represents an ideal vertebrate model system to study embryonic development. All developmental stages, including organogenesis, are clearly visible within the embryo and are described in detail by Kimmel et al. and Haffter et al. (2,3).
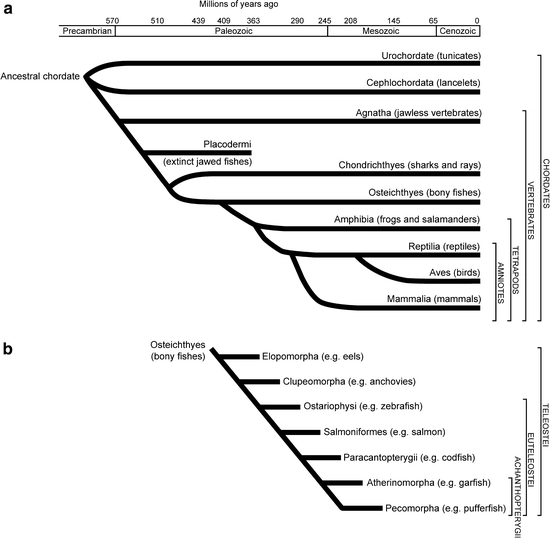

Fig. 1.
Phylogenetic trees of vertebrates and fish.
(a) Evolutionary tree of the chordates and vertebrates. (b) Evolutionary tree of the bony fish (Osteichthyes). Zebrafish belongs to the euteleosti and ostariophysi.
Zebrafish is a relatively simple vertebrate and there is considerable conservation of pathways across species. This means that despite the divergence of teleost fish more than 400 million years ago, they are more closely related to humans than invertebrates and, thereby, offer numerous advantages to researchers interested in many aspects of embryonic development, physiology, and disease. By virtue of their being more closely related to humans, they have many biological functions comparable to human and, hence, many cellular processes have been conserved. Furthermore, it is becoming more and more clear that all vertebrates follow an evolutionary conserved developmental program.
The small size of the zebrafish (3–5 cm), their ability to be kept in large numbers, and the ease of breeding make them easy to maintain. Breeding and collecting eggs from the zebrafish is relatively easy. Since zebrafish are photoperiodic in their breeding, they are kept at a day–night cycle with an automatic timer (14 h light/10 h dark). The day before fertilization, male and female fish are maintained separate using a special breeding tank with two separate compartments divided by a removable partition. By simply removing the partition in the morning, shortly after sunrise, embryos will be produced. As mentioned above, the fertilization of eggs occurs externally and females are very fecund generating hundreds of eggs on a weekly basis.
The embryos develop quickly from a single cell to something that is recognizable as a fish after 24 h of development. Between 4 and 8 h post fertilization (hpf), several characteristic processes occur, including epiboly (migration of cells over the yolk sac), involution, and convergent extension. These processes start with the migration of the cells (approximately 1,200 cells) over the yolk followed by extensive rearrangements. Subsequently, in the next 3 h, the three primary embryonic germ layers are formed through cell movements, a process called gastrulation. After gastrulation is complete, at around 11 hpf, the basic vertebrate body plan has developed and the formation of the first individual somites will begin. Somitogenesis starts anterior (close to the head) and sequentially moves towards the tail of the embryo (posterior). At 18 hpf, 18 somite pairs are formed and the total number that eventually forms is variable, from 30 to 34 pairs. At 24 hpf, a heartbeat and associated blood flow can be recognized. Within 48 hpf, all common vertebrate-specific body features can be seen. Larvae hatch and are able to swim and search for food within 5 days. The generation time of the zebrafish is 2–3 months. For a schematic presentation, see Fig. 2. The external fertilization and the development outside the mother make it easy to access the embryos and to manipulate them, including exposure of larvae/fish to water-soluble chemicals and drugs. The transparency of the embryo enables us to follow the development in vivo. The accessibility and transparency in combination with a fluorescent marker make it possible to visualize detailed developmental cellular movements using real-time imaging. In contrast, the embryonic development of a mouse takes 21 days. To study mouse embryos, the mother has to be sacrificed to get at the embryos, which sacrifices them as well. Owing to these features, the zebrafish has become an important model organism to determine the in vivo function of a gene (functional genomics) during embryonic development.
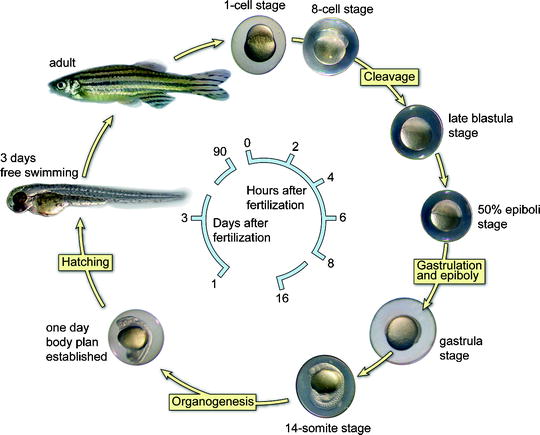

Fig. 2.
A schematic representation of the embryonic developmental stages of zebrafish.
The cycle starts at the top with a fertilized single stage. The embryos develop quickly to a 14-somite stage within 16 h. After 24 h, the complete body plan of the embryo has been established meaning that all organs are present. Two days after fertilization, the embryos hatch and become free swimming. Zebrafish are adult at 3 months of age and fertile.
Two general approaches are used to reveal the function of a gene: forward and reverse genetics. The process of forward genetics starts with a mutant phenotype and moves towards the gene; this was the first approach in functional genomics. The attractive features of zebrafish, as mentioned above, led to large-scale mutagenesis screenings in zebrafish (3–5). It is easy to chemically induce mutations in the germ cells in the testis of adult zebrafish, which may give a phenotype in their offspring. The process of reverse genetics starts with a particular gene and assays the effect of its disruption, that is, studying the phenotype associated with the mutant gene. This approach usually focuses on the inactivation of a gene and requires a way to selectively mutate a chosen gene. In general, the inactivation of the chosen gene is accomplished by homologous gene targeting, resulting in a complete (or sometimes conditional) knockout of the gene. Tools for reverse genetics typically include: gene knockouts/knock-ins using homologous recombination in embryonic stem (ES) cells, gene knock down using morpholino antisense technology (MO; see section gene knock down technologies for detailed information) or RNA Interference (RNAi), and targeted induced local lesions in genomes (TILLING) technology. Some of these applications in zebrafish research will be discussed later in more detail.
Zebrafish models generated by both forward and reverse genetics are not only used as a genetic or developmental model system. In the last decade, zebrafish has been successfully applied as an organism to elucidate the etiology of human disease. Zebrafish models of human disease are widely used in many different fields of medical research, like cancer, infectious diseases, cardiovascular disease, kidney disease, diabetes, blindness, deafness, digestive diseases, hematopoiesis, muscle disorders, and neural disorders (5,6).
In summary, the zebrafish has become a well-established model organism, making important contributions to the identification and characterization of genes and pathways involved in development, organ function, and behavior. Additionally, it has become a valuable resource for identifying genes involved in human disease.
2 The Zebrafish Genome
The zebrafish is a vertebrate with a diploid genome consisting of 25 chromosomes (1n). Although the exact number of genes in zebrafish is currently unknown, estimates about the number of base pairs point to approximately 1.7 × 109 base pairs for the haploid genome. The zebrafish is expected to have at least the same number of genes as the humans. Almost all human genes can be found in the zebrafish and approximately 20% of the human genes have two orthologs in zebrafish. This suggests genome duplication shortly before the teleost radiation, which was either partial or was followed by rapid gene loss. Importantly, if two orthologs are present, they often show different expression patterns (both spatial and temporal). This suggests that the function of the ancestral gene has been divided up between two orthologs with more restricted (less complex) functions (7,8).
Importantly, orthologs of the genes involved in familial Alzheimer’s disease and frontotemporal dementia have been identified in zebrafish as well, including presenilin 1, presenilin 2, APP, Progranulin (GranulinA and GranulinB), and TDP-43 (TAR DNA binding protein of 43 kDa) (9–13).
In February 2001, the Sanger Institute started sequencing the genome of the zebrafish (www.sanger.ac.uk/Projects/D_rerio/). This whole genome-sequencing project leads to the identification of genes of which the in vivo function is unknown. The assembled genomic zebrafish sequences are publicly available (http://www.ensembl.org/Danio_rerio/; http://vega.sanger.ac.uk/Danio_rerio/).
3 Mutagenesis Screens
The zebrafish is very well suitable for large-scale forward genetic screens in which phenotypic defects are identified before the identification of the gene causing these defects. This is due to its large quantity of eggs, short generation time, and the external development of the transparent embryos. In addition, an important practical advantage is that zebrafish sperm can be frozen for future studies. The three techniques to induce mutations in zebrafish include chemical mutagenesis, gamma irradiation, and insertional mutagenesis (5).
Chemical mutagenesis is the most favorable and efficient method applied thus far (3,14). The chemical mutagenesis by exposing adult zebrafish to N-ethyl-N-nitrosourea (ENU) is used for these screens. ENU, an alkylating agent, generates point mutations throughout the entire genome in premeiotic germ cells by transferring its ethyl group to individual bases of the DNA, which are misread by DNA polymerase in subsequent replications. However, most induced mutations are recessive and must be rendered to homozygosity to reveal a phenotype. This is accomplished by a multigeneration backcross model (3). Mutagenized adult male zebrafish are crossed with wild-type females. The F1 offspring are heterozygous for individual mutations and are once more crossed with wild-type females. The resulting F2 generation is intercrossed randomly to produce F3 families in which homozygous mutations occur. In the F1 and F2 generation, rare dominant mutations might occur, but most ENU-induced mutations are recessive. Finally, the F3 embryos are further selected on defects in organogenesis using microscopic examination between 1 and 5 days post fertilization (dpf). Positional cloning can then identify the affected gene in isolated mutants. Large numbers of mutations that disrupt embryonic development have now been isolated in the zebrafish, many of which may serve as models for human diseases or syndromes. Further characterization of these mutants (∼2,000) and identification of the genetic defect will advance our knowledge of the pathogenesis of the corresponding human disease. It will advance our understanding of the underlying molecular basis of the disease and ultimately may lead to the development of drugs aimed at treatment of the disease.
The second approach to induce mutations disrupting developmental processes in zebrafish is radiation, mainly gamma. Gamma-ray mutagenesis produces a very high locus mutation rate of approximately 1:100 and has mainly been used in screens for morphological defects (2). In contrast to chemical mutagenesis, gamma rays induce translocations and large deletions at high frequency in the zebrafish genome and thus chemical mutagenesis is the method of first choice (15).
The third alternative approach to induce mutations in zebrafish is insertional mutagenesis, which can be established by injection of plasmid DNA, a mouse pseudotyped retrovirus or using a P-element transposon as insertional mutagens (16). For retroviruses, a molecular tag at the site of the mutagenic lesion enables detection of the mutated gene. Although the efficiency of the mutagenesis is less than with ENU mutagenesis, the detection is seven to eight times higher than for ENU-induced mutations (17,18). These genetic screens allow the identification of novel genes and mutants for specific organs or processes. After examining the phenotypes by random mutagenesis, the mutation responsible for the specific defect has to be found using positional cloning. Major drawback of forward genetics is that it is slow and laborious due to positional cloning methods.
Mutagenic screening technology using a reverse genetic approach has been established as well. In 2002, owing to the lack of a working protocol to produce an ES-cell-based knockout (or targeted gene expression), the TILLING technique was developed (19–21). TILLING involves randomly induced mutations by ENU and subsequent screening for mutations in target genes. This screen is an enzyme-mediated (CEL-I endonuclease) mismatch recognition procedure to detect heterozygous germline mutations in the F1 generation. Further generation of embryos with mutant phenotypes is similar to the breeding scheme described above. TILLING can be performed in a high-throughput setup. The only disadvantage is that the mutations are randomly introduced. The TILLING method would lead to more null mutants in zebrafish than achieved by homologous recombination in mice. Initially, TILLING was developed to reduce the time and costs of mutation detection using DNA sequencing. However, recent advances in DNA sequencing technology make this method equal to TILLING.
4 Transgenic and Knockout Zebrafish
Genetically modified animal models are widely used to characterize the function of many newly identified (disease) genes. Transgenic techniques in the mouse to generate transgenic and loss-of-function mutations are well established and have significantly improved our understanding of the roles of specific gene products. Genetically modified mice also serve as valuable models to study the pathogenesis of human disease and to test or develop experimental treatment regimes. However, with the zebrafish emerging as an important model organism to study human disease, the development of similar or additional genetic techniques specifically focused on zebrafish was needed. Methods for generating a transgenic zebrafish are pseudotyped retrovirus infection (17,22,23), transposons (24–26), transfection of sperm nuclei (27), and DNA microinjection. The latter is the most frequently used method for generating transgenic lines expressing a gene of interest. DNA microinjection can be achieved by injection of plasmid DNA or bacterial artificial clones (BACs) into the cytoplasm of a 1-cell stage embryo. The frequency of DNA integration into the germline by microinjection in zebrafish is 1–30%, which is comparable to mouse (28). Coinjection of I-SceI meganuclease and a construct flanked by meganuclease recognition sites has been shown to improve the integration in fish (29). The gene of interest, often cloned in a fusion vector containing a cDNA encoding Enhanced Green Fluorescent Protein (EGFP), is randomly integrated or under the control of a general or tissue-specific promoter. This approach has been successfully applied in the generation of a transgenic zebrafish model for dementia (see later).
Another approach to investigate the function of a (disease) gene in an animal model is to inactivate or disrupt the gene of interest. A major advancement in the ability to generate mouse disease models was the development of technology that makes it possible to introduce loss-of-function mutations into endogenous genes and then transmit these through the mouse germline (30). The desired null mutations are first created via homologous recombination in ES cells, which contribute to all cell lineages when injected into blastocysts. However, mouse developmental genetics is impeded by the high cost of maintenance of animals and by the intrauterine mode of development. Because of the expense and effort required to produce a genetically modified mouse, and the inaccessibility of the embryos inside the mother, the zebrafish might be the vertebrate model to allow these genetic techniques.
The strategy to generate knockout zebrafish by the germline transmission of targeted loss-of-function alleles using ES cells, as described for mice, has not yet been achieved. The only method to produce a knockout zebrafish was mutagenesis followed by targeted screening for point mutations as described earlier (TILLING) (19). Pluripotent zebrafish ES cell lines have been established (31). Recently, targeted incorporation of plasmid DNA into these cells by homologous recombination followed by in vitro drug selection was successful (32). In addition, the authors were able to introduce these ES cells, expressing a marker gene such as EGFP, into host embryos using microinjection techniques and achieved contribution to the germline. Although the frequency of germline chimera production was 2–4%, the availability of large quantities of fertilized eggs makes it potentially feasible to establish a knockout fish line in the near future (32).
5 Gene Knock Down Technologies
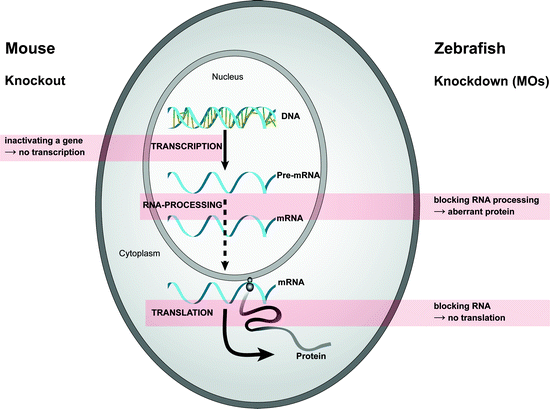
Zebrafish reverse genetics is slowly catching up with Drosophila and/or mouse, as the techniques to perform gene-specific knock downs, target-selected mutagenesis, and transgenesis in zebrafish are quickly developing. Next to the ability to make a knockout (see above), the generation of a morpholino-mediated knock down zebrafish was the favorite technology thus far to study gene function in zebrafish (6,33). Currently, the use of antisense modified oligonucleotides is still widely applicable due to their ease and quick results. Although morpholinos have been tested in different species, including sea urchin, ascidian, frog, chicken, and mouse, the most favorable model organism to test morpholinos has been carried out on zebrafish embryos. MOs are synthetic oligonucleotides of 25 bases, which hybridize specifically to complementary sequences of mRNA, thereby disrupting translation initiation or pre-mRNA splicing. The backbone of MOs is similar to the backbone of DNA or RNA, but with some changes. In MOs, the ribose or deoxyribose sugar molecules that link the bases of the DNA or RNA are replaced by morpholino rings (hence the name). Anionic phosphates of bases replace nonionic phosphorodiamidate linkages. Because of this modified backbone, MOs are uncharged, very stable, and cannot be degraded by nucleases (for more details about MOs, see www.gene-tools.com). The reduction of translation will never be 100%, but can be up to 90%, and is therefore called a knock down (Fig. 3). Zebrafish embryos displaying a phenotype as a consequence of ectopic MO administration are called morphants (34).
 < div class='tao-gold-member'>
< div class='tao-gold-member'>





Only gold members can continue reading. Log In or Register to continue
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


