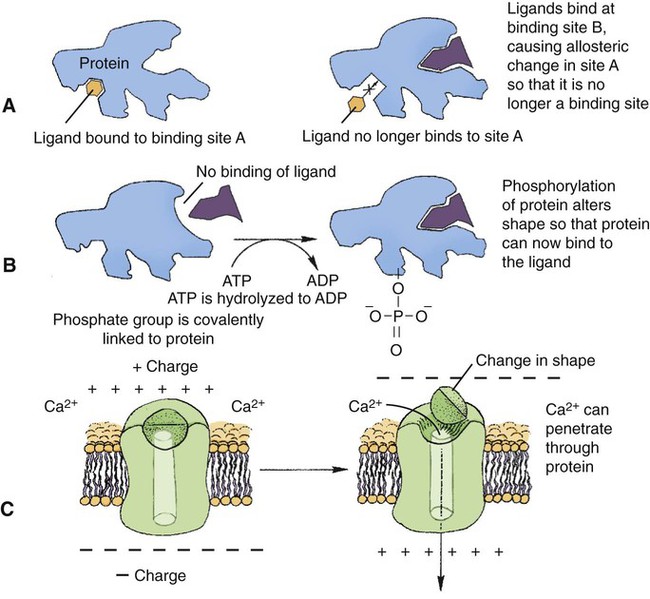
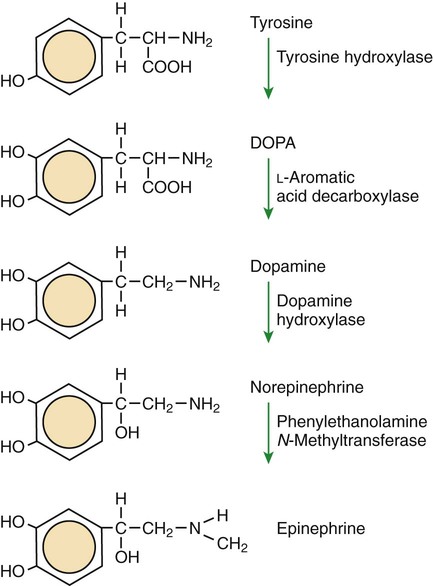
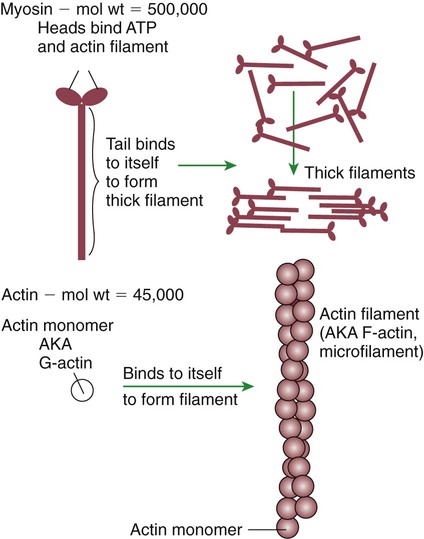
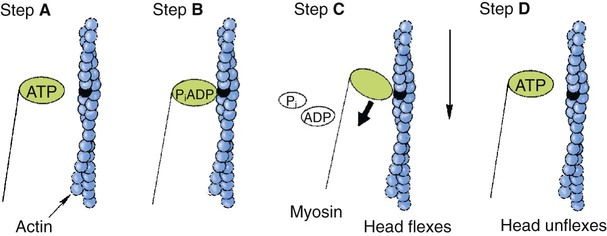
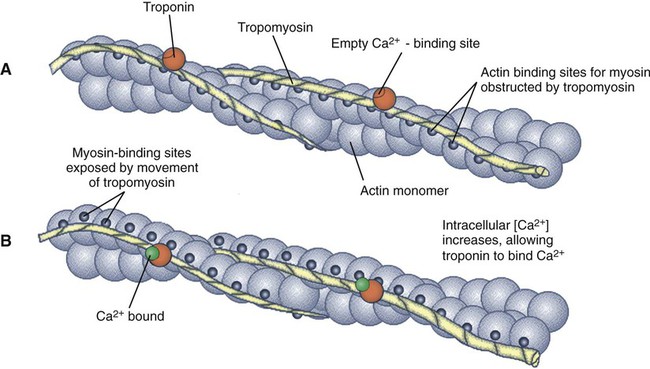
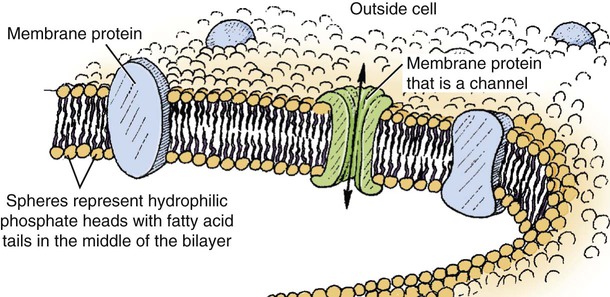
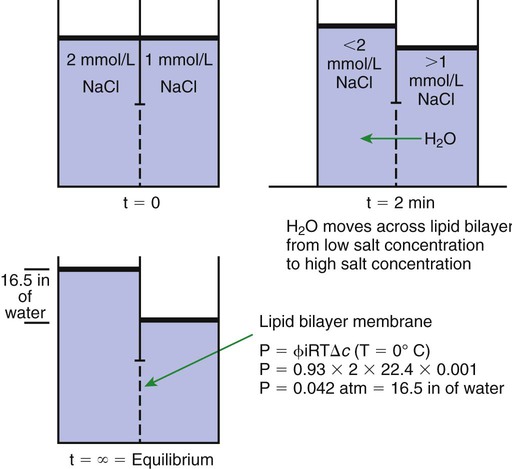
1. All physiological change is mediated by proteins. 2. Protein function depends on protein shape and shape changes. 3. A series of enzymatic reactions converts tyrosine into the signaling molecules dopamine, norepinephrine, and epinephrine. 4. Muscle contraction and its initiation and cessation depend on the binding specificity and allosteric properties of proteins. 5. Biological membranes are a mosaic of proteins embedded in a phospholipid bilayer. 1. Only small, uncharged molecules and oily molecules can penetrate biomembranes without the aid of proteins. 2. Molecules move spontaneously from regions of high free energy to regions of lower free energy. 3. Important transport equations summarize the contributions of the various driving forces. 4. Starling’s hypothesis relates fluid flow across the capillaries to hydrostatic pressure and osmotic pressure. 5. Membrane proteins that serve the triple functions of selective transport, catalysis, and coupling can pump ions and molecules to regions of higher free energy. 6. Many membrane proteins selectively facilitate the transport of ions/molecules from high to low electrochemical potential. 7. Passive transport of K+ across the plasma membrane creates an electrical potential. 8. Spatial organization of active and passive transport proteins enables material to pass completely through the cell. 9. Membrane fusion allows for a combination of compartmentalization and transport of material. Information transmission and transduction 1. Cell signaling often occurs by a lengthy chain of sequential molecular interactions. 2. Signaling pathways begin with the binding of an extracellular molecule to a receptor. 3. Specific physiological information is inherent in the receptor/ligand complex, not in the hormone/neurotransmitter molecule. 4. G-protein–coupled receptors are the largest family (a superfamily) of receptors and help regulate almost all physiological processes. 5. Most G-protein–linked information is sent to the cytoplasm by second messengers. 6. Ca2+ transport across plasma and intracellular membranes is an important second messenger. 7. Cyclic AMP is produced by activation of a membrane-bound enzyme in response to hormone/neurotransmitter binding to receptors. 8. The receptor-mediated hydrolysis of a rare phospholipid of the plasma membrane produces two different second messengers with different actions. 9. Steroid hormones and other lipid signals interact with nuclear receptors, which are transcription factors within the cell. Allosteric changes in protein conformation arise in four general ways. One way, just mentioned, is that most proteins change shape depending on which ligands are bound at particular binding sites (Figure 1-1, A). The sequence—specific ligand binding → protein shape change → change in protein-binding properties and protein function → this change regulates something—is a common molecular mechanism underlying physiological control. This method involves no alteration in the covalent structure of the protein. A second method of producing conformational change, however, occurs as a result of the covalent modification of one or more of the amino acid side groups of the protein (see Figure 1-1, B). By far the most common such change is the covalent addition of a phosphate group to the hydroxyl (—OH) group on the side chain of serine, threonine, or tyrosine residues in the protein. This modification is called phosphorylation. Because the phosphate group is highly charged, phosphorylation of a protein alters hydrogen bonding and other electrostatic interactions within the protein chain, altering its conformation and functional properties. In a third method, some physiologically important proteins change shape in response to the electrical field surrounding the protein (see Figure 1-1, C). These respond to a voltage change by altering the position of charged amino acids, thus altering protein shape. Figure 1-2 is a diagram of the series of reactions by which the amino acid tyrosine is converted into three different signaling molecules: (1) dopamine, a brain neurotransmitter; (2) norepinephrine, a neurotransmitter of the brain and peripheral autonomic nervous system; and (3) epinephrine, an autonomic neurotransmitter and hormone. Dopamine, norepinephrine, and epinephrine share a similar structure. All contain a phenyl (benzene) ring with two hydroxyl groups (i.e., catechol) and an amine group (thus catecholamines). They are among the large number of molecules that function as neurotransmitters. That is, the electrically coded information sent along nerve cells causes the release of a chemical, the neurotransmitter, at the terminal of the neuron, which is next to a target cell, such as another nerve, a muscle, or an endocrine cell. The electrically encoded information of the nerve is transmitted to the target cell by the binding of the neurotransmitter to proteins on the surface of the target cell. Proper neurotransmitter synthesis is crucial to nervous function and physiological regulation. Myosin is a large protein whose shape resembles a two-headed golf club. The elongated tail of the myosin molecule corresponds to the shaft of the golf club, and there are two knobs at one end of the tail that, as with golf clubs, are called heads. Myosin tails bind specifically to other myosin tails, forming bipolar aggregates called thick filaments (Figure 1-3). Myosin heads specifically bind ATP and another muscle protein, actin. Actin binds to itself to form long, thin filaments, called thin filaments in muscle and called F-actin (filamentous actin) in other cell types. Actin filaments play an important architectural role in all animal cells. Although actin is best understood in muscle cells, all animal cells depend on actin filaments for their shape and for their capacity to migrate in their environment. Actin filaments can be “woven” in various ways to produce different structures, such as ropelike bundles and clothlike networks. These actin bundles and actin networks are used to support the cell in particular shapes, similar to ropes holding up the woven cloth of a tent. In muscle, the interaction of myosin, ATP, and actin produces contraction and force, as shown in Figure 1-4: Step A: ATP binds to a myosin head; in this conformation, myosin has little ability to bind to actin. Step B: Enzymatic activity associated with the myosin head, an adenosinetriphosphatase (ATPase), rapidly causes a partial hydrolysis of ATP to adenosine diphosphate (ADP) and inorganic phosphate (Pi), both of which stay bound to the myosin. With ADP and Pi bound, myosin has a slightly different shape that binds avidly to nearby actin filaments. Step C: When myosin binds to actin, called cross-bridging, the myosin head couples the complete hydrolysis of ATP to a forceful flexing of the myosin head. This allosteric change causes the actin filament to slide past the thick filament. This sliding puts the actin filament under tension, which in turn causes the muscle to contract (shorten) against the load of the muscle (i.e., lifting a weight or pumping out blood). All muscle contraction depends on this sliding filament mechanism of actin and myosin-based filaments. This same allosteric change of myosin also alters myosin-binding properties so that it releases the ADP and Pi. Step D: The binding of a new ATP molecule to the myosin head again causes myosin to change shape; the head unflexes and loses its affinity for actin, releasing the cross-bridge, and the cycle can start over. Rigor mortis of dead animals is caused by a lack of new ATP to bind to myosin heads. In the absence of ATP, myosin heads remain in Step C (i.e., bound to actin). The muscle is stiff because it is completely cross-bridged together. This system of contractile proteins requires some control so that, for example, the heart beats rhythmically and skeletal muscle contraction is coordinated. At the organismal level, skeletal and cardiac muscle contraction is primarily under control by electrical stimulation from nerves or other electrically active cells (see Chapter 6). The transmission of electrical excitation to the actomyosin system is called excitation-contraction coupling. Excitation-contraction coupling in all types of muscle depends on changes in intracellular calcium ion (Ca2+) concentration. In skeletal and cardiac muscle, but not smooth muscle, two additional thin-filament proteins, troponin and tropomyosin, are required for this coupling. (Excitation-contraction coupling for smooth muscle is discussed later in this chapter.) In striated muscles, troponin binds to tropomyosin and to Ca2+. Tropomyosin is a long, thin protein that binds in the groove of the actin filament in such a way that its positions, high in the groove or snuggled down deep in the groove, allow or prevent the myosin head access to the thin filament (Figure 1-5). Excitation-contraction coupling of striated muscle works as follows: Step A: Electrical excitation of a striated muscle cell causes an increase in the intracellular concentration of Ca2+. Step B: The additional Ca2+ binds to troponin, causing an allosteric change in troponin. Step C: Because Ca2+ is bound to troponin, which in turn is bound to tropomyosin, the Ca2+-induced change in troponin conformation is transmitted to the tropomyosin molecule. When troponin binds Ca2+, tropomyosin changes its binding to actin in such a way that it exposes the actin site for myosin cross-bridging. (Tropomyosin snuggles down deeper in its actin groove, revealing actin to the myosin head.) As long as troponin binds Ca2+, the muscle contracts by the actomyosin cycle outlined earlier. Step D: When the Ca2+ concentration drops to normal, however, troponin no longer binds Ca2+. This causes tropomyosin to move up in the thin filament groove so that it again blocks the myosin-binding sites on actin. Myosin heads can no longer cross-bridge, and muscle contraction stops. Before continuing the discussion of the cellular basis of physiological control, an additional basic structure must be introduced. This is the phospholipid bilayer of the biomembranes of cells. Phospholipids are molecules that have two long tails of hydrophobic fatty acid and a head containing a charged, hydrophilic phosphate group. Under appropriate aqueous conditions, these molecules spontaneously form an organized membrane structure, similar to the film of a soap bubble. This filmy layer is composed of two layers (a bilayer) of phospholipid molecules. In both layers the hydrophilic heads point outward to hydrogen bond with water, and the oily, fatty-acid tails point inward, toward one another and away from the water. Proteins embedded in this lipid bilayer, called intrinsic membrane proteins or just membrane proteins, produce the fluid mosaic structure of biomembranes shown in Figure 1-6. All biological membranes share this fluid mosaic structure, whether the membrane is the outer plasma membrane separating cytoplasm from extracellular fluid or the membrane surrounding intracellular membranous organelles such as endoplasmic reticulum or lysosomes. It is called a fluid mosaic because of the mosaic of proteins among phospholipids, and because the phospholipid layer is fluid; proteins can move around and diffuse within the plane of the bilayer “like icebergs floating in a phospholipid sea” (the apt phrase of S. J. Singer, one of the originators of the model). Charged particles (i.e., ions) do not pass through a pure phospholipid bilayer because of the inner, hydrophobic region of bilayer. Polar molecules (molecules with no net charge but with electrical imbalances) with a molecular weight greater than about 100 daltons are also unable to pass readily through a pure lipid bilayer, thus excluding all sugar molecules (monosaccharides), amino acids, nucleosides, as well as their polymers (polysaccharide, proteins, nucleic acids). On the other hand, some crucially important polar molecules (e.g., water, urea) are small enough to pass through the lipid bilayer. Small, moderate-size, and large molecules that are soluble in oily solvents readily pass through a pure lipid bilayer. Physiologically important molecules in this class include O2, N2, and the steroid hormones (see Chapters 33 and 34). However, many toxic, synthetic molecules, such as insecticides, are also in this category. One of these equations relates a hydrostatic (pressure) driving force for water movement that just balances a driving force caused by a chemical potential difference. Osmosis is the movement of water across a semipermeable membrane in response to the difference in the electrochemical potential of water on the two sides of the membrane (Figure 1-7). The chemical potential of water is lower in 1 liter (L) of water (H2O) in which is dissolved 2 millimoles (mmol) of sodium chloride (NaCl) than in 1 L of H2O in which is dissolved 1 mmol of NaCl. If these two solutions are separated by a pure lipid bilayer, Na+ and Cl– ions cannot move to equilibrate the concentration. Rather, the freely permeable water moves from the side with the higher water potential (low concentration of solute) to the side with the lower water potential (higher concentration of solute). Thus, water follows solute (a good summary of osmosis), and this water movement dilutes the 2 mmol solution. However, water movement never produces equal concentrations of salt. Rather, another driving force appears as the water moves. The hydrostatic pressure of water increases on the side to which the water moves, increasing the electrochemical potential of the water on that side. Net water movement stops when the increase in water potential from hydrostatic pressure exactly balances the decrease in water potential from the dissolved salt, so that the electrochemical potential becomes equal on both sides of the membrane. The initial potential difference of water shown in Figure 1-7 is caused by the difference in the concentration of material dissolved in the water. A proper explanation of why the water in a solution has a lower chemical potential than pure water (and why water in a concentrated solution has a lower potential than in a dilute solution) is beyond the scope of this chapter. However, readers familiar with the concept of entropy will realize that the disorder of a system increases with the introduction of different particles into a pure substance and with the number of different particles introduced. An analogy would be that a canister with mixed sugar and salt is more disordered, and therefore at higher entropy, than a canister with only pure salt or pure sugar. Also, the disorder of the system increases as more sugar is added to salt (up to 50 : 50); a pinch of sugar in a canister of salt only increases the disorder slightly. Because an increase in entropy causes a decrease in free energy, the free energy of a solution is decreased as the mole fraction of solute increases. Π =Osmotic pressure, the driving force for water movement expressed as an equivalent hydrostatic pressure in atmospheres (1 atm = 15.2 lb/in2 = 760 mm Hg). Osmotic pressure is symbolized by Π to distinguish it from other types of pressure terms. i =Number of ions formed by dissociating solutes (e.g., 2 for NaCl, 3 for CaCl2). R =Gas constant = 0.082 L atm/mol degree. T =Temperature on the Kelvin scale; 0° C = 273° K. Δc =Difference in the molar concentration of the impermeable substance across the membrane.
The Molecular and Cellular Bases of Physiological Regulation
Protein Function Depends on Protein Shape and Shape Changes
A Series of Enzymatic Reactions Converts Tyrosine into the Signaling Molecules Dopamine, Norepinephrine, and Epinephrine
Muscle Contraction and its Initiation and Cessation Depend on the Binding Specificity and Allosteric Properties of Proteins
Biological Membranes Are a Mosaic of Proteins Embedded in a Phospholipid Bilayer
Transport
Only Small, Uncharged Molecules and Oily Molecules Can Penetrate Biomembranes Without the Aid of Proteins
Important Transport Equations Summarize the Contributions of the Various Driving Forces
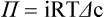
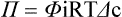
Starling’s Hypothesis Relates Fluid Flow Across the Capillaries to Hydrostatic Pressure and Osmotic Pressure
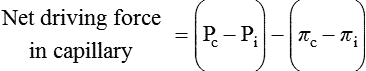
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


The Molecular and Cellular Bases of Physiological Regulation
Only gold members can continue reading. Log In or Register to continue