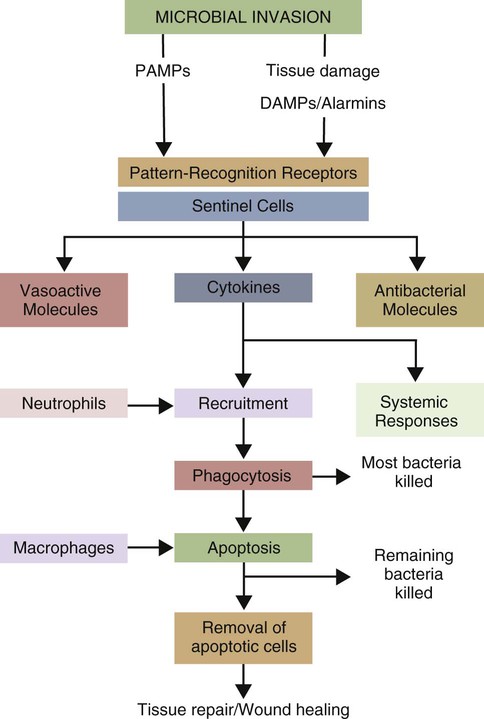
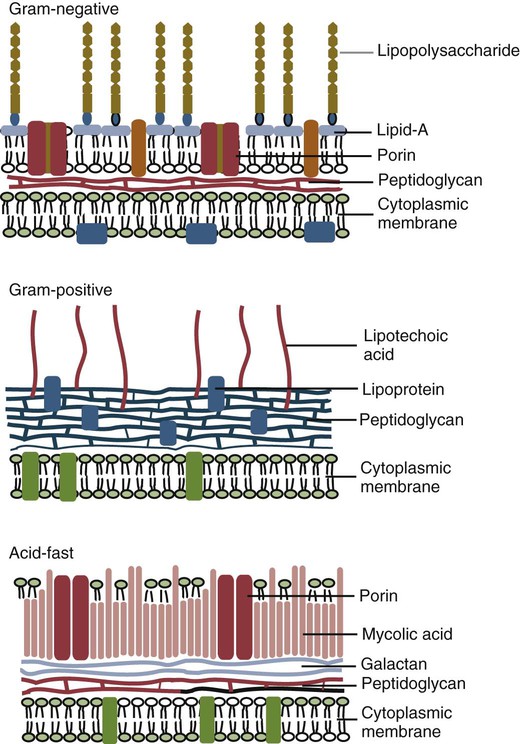
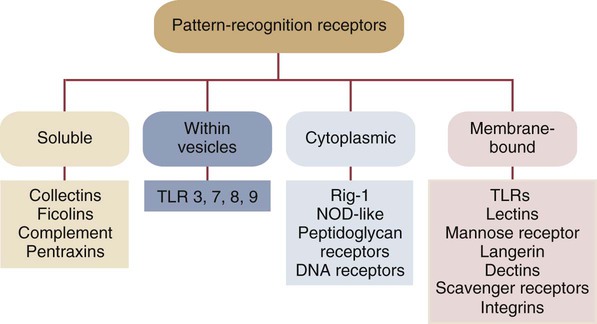
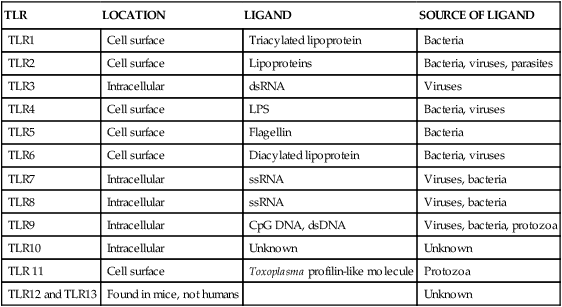
• Two types of signal trigger the body’s innate defenses. One signal generated by the presence of invading microorganisms is detected by sensing their characteristic surface molecules, or nucleic acids. These are called pathogen-associated molecular patterns (PAMPs). • Cells also detect molecules released from damaged tissues and broken cells. These are called damage-associated molecular patterns (DAMPs) or alarmins. • Both PAMPs and DAMPs bind to pattern-recognition receptors (PRRs) on cell surfaces or located within cells. • PRRs are found on many different cell types. The most important of these “sentinel” cells are macrophages, dendritic cells, and mast cells. • A major group of PRRs is called toll-like receptors (TLRs). • Signals generated when PAMPs bind TLRs activate sentinel cells and stimulate them to secrete many different molecules. Some of these molecules are proteins called cytokines that “turn on” the inflammatory process. • These molecules trigger local increases in blood flow, attract defensive cells such as neutrophils, and increase blood vessel permeability, allowing antimicrobial molecules and cells to flood affected tissues. Inflammation concentrates defensive cells and antimicrobial molecules at sites of microbial invasion and tissue damage. These defensive cells are the white blood cells (leukocytes) that circulate constantly in the bloodstream. Inflammation triggers the migration of leukocytes, from the bloodstream to sites of invasion where they attack and destroy invaders. Likewise, many protective proteins, such as antibodies and complement components, are normally found only in blood and can only enter tissues during inflammation. Inflammation is therefore a mechanism by which defensive cells and proteins are focused on sites of microbial invasion. Together, they destroy the invaders and then repair any subsequent tissue damage (Figure 2-1). Microbes not only grow very fast but also are highly diverse and can mutate and change many of their surface molecules very rapidly. For this reason, the innate immune system does not attempt to recognize all possible microbial molecules. Rather, the body uses receptors that can bind and respond to abundant, essential molecules that are common to many different microorganisms but are absent from normal animal tissues. Because they play essential roles, they tend not to change rapidly. They are essential for microbial survival, and as a result, are commonly shared by entire classes of pathogens. They are, in effect, widely distributed molecular patterns. For example, the walls of Gram-positive bacteria are largely composed of peptidoglycans (chains of alternating N-acetylglucosamine and N-acetylmuramic acid cross-linked by short peptide side chains) (Figure 2-2). Gram-positive bacterial cell walls also contain lipoteichoic acids. The cell walls of Gram-negative bacteria consist of peptidoglycans covered by a layer of lipopolysaccharide (LPS). Acid-fast bacteria are covered in glycolipids. Yeasts have a mannan- or β-glucan–rich cell wall. Viruses, in contrast, grow within infected host cells, so the main targets of antiviral PRRs are unique viral nucleic acids. Many different PRRs are used to ensure as complete coverage as possible of PAMPs. Most are cell-associated receptors found on cell membranes, within the cytosol, and within cytoplasmic vesicles. Soluble receptors also circulate in the bloodstream (Figure 2-3). Examples of PRRs include the toll-like receptors (TLRs), the retinoic acid-inducible gene (RIG)-1-like receptors (RLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), and the C-type lectin receptors (CLRs). CLRs are soluble proteins primarily involved in the capture and uptake of invading bacteria. The others, in contrast, are receptors that activate intracellular signaling cascades. The most important family of PRRs consists of the TLRs (Box 2-1). Some TLRs are located on cell surfaces, where they are responsible for recognizing extracellular invaders such as bacteria and fungi. Other TLRs are located within the cells, where they are responsible for detecting intracellular invaders such as viruses. TLRs are a critical first line of defense against bacterial, viral, and fungal invaders and play a vital role in microbial sensing. TLRs are transmembrane glycoprotein receptors. Mammals possess 10 or 12 different functional TLRs (TLR1 to TLR10 in humans and cattle and TLR1 to TLR9 and TLR11 to TLR13 in mice) (Table 2-1). The cell surface TLRs (TLR1, 2, 4, 5, 6, and 11) mainly recognize bacterial and fungal proteins, lipoproteins, and lipopolysaccharides. The intracellular TLRs (TLR3, 7, 8, 9, and 10), in contrast, recognize viral and bacterial nucleic acids. For example, TLR4 on the cell surface recognizes lipopolysaccharides on Gram-negative bacteria. TLR2 recognizes peptidoglycans, lipoproteins, and a glycolipid called lipoarabinomannan from Mycobacterium tuberculosis, whereas TLR5 recognizes flagellin, the major protein of bacterial flagella. TLR9 is an intracellular sensor of bacterial DNA and is triggered by intracellular bacteria. Other intracellular receptors, such as TLR3 and TLR7, recognize viral double-stranded (ds) ribonucleic acid (RNA), whereas TLR7 and TLR8 recognize viral single-stranded (ss) RNA. Most TLRs are usually homodimers formed by identical paired peptide chains. They may also form heterodimers using dissimilar chains. For example, one TLR2 chain can associate with a TLR6 chain, and this heterodimer can then recognize bacterial diacylated lipopeptides. A TLR2 chain also associates with a TLR1 chain to recognize mycobacterial triacylated lipopeptides. Given the number of possible TLR chain combinations, it is believed that the presently known TLRs can collectively recognize almost all PAMPs. TLR11 is different from the other TLRs. It is found only on dendritic cells, macrophages, and epithelial cells in the mouse urinary tract, where it recognizes bacteria and PAMPs from protozoan parasites (Box 2-2). Table 2-1 Pathogen-Associated Molecular Patterns Recognized by the Mammalian Toll-like Receptors CpG, cytosine-guanosine; LPS, lipopolysaccharide; TLR, toll-like receptor. When a PAMP binds to its corresponding TLR, signals are passed to the cell. Multiprotein signaling complexes are formed, signal transduction cascades are initiated, and as a result, proinflammatory molecules are produced by the cell. Each step in the process involves multiple biochemical reactions involving many different proteins, and the precise signaling pathway employed differs between TLRs. For example, the cell surface TLRs use different pathways than the intracellular TLRs. All TLRs except TLR3 use an adaptor protein called MyD88 to activate three major transcription factors, nuclear factor kappa-B (NF-κB), MAP kinase (MAPK), and IRF3 (Figure 2-4). NF-κB and MAPK activate the genes for three major proteins, interleukin-1 (IL-1), interleukin-6 (IL-6), and tumor necrosis factor-alpha (TNF-α) IRF3 activates the gene for another protein, interferon-β. (For additional details on signal transduction, see Chapter 8.) The proteins produced by these sentinel cells are all classed as cytokines, proteins that regulate the activities of cells involved in the defense of the body. The cytokines are produced as inactive precursors and then activated by an enzyme called caspase-1. The production of caspase-1 is triggered by a protein complex called an inflammasome (Box 2-3).
Innate Immunity
The Recognition of Invaders
How Invaders Are Recognized
Pathogen-Associated Molecular Patterns
Toll-like Receptors
TLR
LOCATION
LIGAND
SOURCE OF LIGAND
TLR1
Cell surface
Triacylated lipoprotein
Bacteria
TLR2
Cell surface
Lipoproteins
Bacteria, viruses, parasites
TLR3
Intracellular
dsRNA
Viruses
TLR4
Cell surface
LPS
Bacteria, viruses
TLR5
Cell surface
Flagellin
Bacteria
TLR6
Cell surface
Diacylated lipoprotein
Bacteria, viruses
TLR7
Intracellular
ssRNA
Viruses, bacteria
TLR8
Intracellular
ssRNA
Viruses, bacteria
TLR9
Intracellular
CpG DNA, dsDNA
Viruses, bacteria, protozoa
TLR10
Intracellular
Unknown
Unknown
TLR 11
Cell surface
Toxoplasma profilin-like molecule
Protozoa
TLR12 and TLR13
Found in mice, not humans
Unknown
< div class='tao-gold-member'>
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Innate Immunity: The Recognition of Invaders
Only gold members can continue reading. Log In or Register to continue