Fig. 1
Schematic diagram of a colorimetric assay based on unmodified AuNPs for detection of unamplified nucleic acids (SVCV RNA). First, the target RNA is denatured and the complementary probe hybridizes to the target forming double strands. Adding AuNPs causes its aggregation since the probe is not free to stabilize the AuNPs, and the solution color changes from red to blue. In the presence of a noncomplementary target RNA, the probe will be free to adsorb onto and stabilize the AuNPs consequently preventing their aggregation and the solution color remains red
2 Materials
2.1 Gold Nanoparticles
1.
38.8 mM sodium citrate solution (Sigma-Aldrich, GmbH, Schnelldorf, Germany).
2.
1 mM aqueous solution of gold (III) chloride trihydrate (HAuCl4) (Sigma-Aldrich, GmbH, Schnelldorf, Germany).
2.2 Oligonucleotide Probes
1.
Oligonucleotide probes targeting specific sequences of a particular gene of interest of the pathogen. For example, we have recently described the development of a 26 bp specific probe (5′-GTC TAT CAT CAG CTA CAT CGC ATT CC-3′) designed to target the spring viremia of carp virus (SVCV) glycoprotein gene [29]. Assess the specificity of the probe against other common aquatic pathogens sequences deposited in GenBank.
2.
Stock solutions of the probes: order the designed probes commercially and prepare them according to the supplied specification sheet that usually gives instructions needed to rehydrate the probes. For each probe, add the recommended amount of purified, double-distilled, deionized, and autoclaved water (PCR grade water), and mix well to get a stock solution of 100 pmol/μl. Prepare several aliquots from these stock solutions to avoid degradation by repeated freezing and thawing, and keep at −20 °C until required.
2.3 Colorimetric Detection of Nucleic Acids
1.
2.
Hybridization buffer: 10 mM phosphate buffer saline (PBS), pH 7.0, containing 0.4 M of NaCl and 1.8 μM of oligonucleotide probe (see Note 2 ).
3 Methods
3.1 Preparation of Gold Nanoparticles
Prepare gold nanoparticles of 13 nm diameter by the citrate reduction method according to Grabar et al. [32] or purchase commercially (e.g., from Strem Chemicals. Newburyport, MA, USA).
1.
Add 10 ml of 38.8 mM sodium citrate solution rapidly to 100 ml of vigorously stirred boiling 1 mM HAuCl4 aqueous solution (see Note 3 ).
2.
The mixture should be boiled for 10 min and stirred for an additional 15 min.
3.
Allow the solution to cool to room temperature.
4.
Filter and store at 4 °C before use.
3.2 Colorimetric Detection of Nucleic Acids Using Unmodified Gold Nanoparticles
Optimum concentrations of NaCl and oligonucleotide probe (see Note 1 ), as well as optimal pH, annealing temperature, and incubation times, need to be determined for each assay. We give an example based on the detection of unamplified SVCV RNA that we recently described [29].
The assay is performed as follows:
1.
In a sterile PCR tube place 5 μl of extracted RNA.
2.
Add 3 μl of the hybridization buffer.
3.
Complete the reaction mixture to 10 μl with sterile distilled water and mix well.
4.
Denature the mixture at 95 °C for 30 s, anneal at 58 °C for 30 s, and then cool at room temperature for 10 min.
5.
Finally, add 10 μl of colloidal AuNPs to the reaction mixture, and observe the change in the solution color within 1 min (Fig. 2) (see Notes 4 – 6 ).
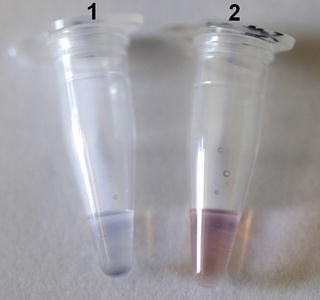

Fig. 2
Colorimetric assay using unmodified AuNPs. Each tube contains 5 μL of sample (SVCV RNA), 1.8 μM of oligonucleotide probe, and 0.1 M of NaCl. The samples were denatured at 95 °C for 30 s and annealed at 58 °C for 30 s and then cooled at room temperature for 10 min. 10 μL of AuNPs solution was added and results were observed within 1 min. Tube 1 contains a positive sample and tube 2 contains a negative sample. In the positive sample, the color changes from red to blue
4 Notes
1.
Inoculate SVCV onto EPC cell line maintained in Eagle’s minimal essential medium (EMEM) buffered to pH 7.6 with sodium bicarbonate, supplemented with 2 % of fetal bovine serum (FBS) and standard concentrations of antibiotic. Incubate the inoculated cultures at 15 °C.
2.




Optimum concentrations of NaCl and of oligonucleotide probe need to be determined for each assay, allowing the visual detection of the color change of the solutions and, at the same time, an effective annealing of the probe to its complementary target. For the detection of SVCV, the concentration of NaCl sufficient for both aggregation of AuNPs and proper annealing of the probe to its complementary target was 0.4 M. Use different concentrations of the probe to determine the optimum probe concentration sufficient to stabilize the AuNPs in the presence of salt. We found that a final probe concentration of more than 3 μM was too high for any aggregation to occur in the presence of the target, leading to false negative results. In contrast, a probe concentration of less than 0.2 μM did not prevent aggregation of AuNPs in the absence of the target and led to false-positive results.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


