and Ellen A. A. Nollen1
(1)
Department of Genetics, University Medical Centre Groningen, University of Groningen, Groningen, The Netherlands
Abstract
The free living nematode worm Caenorhabditis elegans (C. elegans) has been extensively studied by biological, agricultural, and medical scientists for over 40 years. The animal has several characteristics that make it useful as a model organism. For example, the nematodes are transparent, which allows study of embryonic development and gene expression in living animals under the microscope. They also have a very short life cycle of about 3 days and a relatively short lifespan of about 3 weeks, which allow genetic dissection of the mechanisms that affect aging and ultimately determine lifespan. In addition, the mechanism of gene silencing by RNA interference has been discovered in C. elegans and has been developed into a potent reverse genetic tool.
Because of the strong conservation of molecular genetic pathways between C. elegans and mammals, it represents a powerful addition to the small animal model repertoire. Genetic mechanisms in human disease, such as Alzheimer’s disease, have been elucidated in C. elegans, indicating its potential as a model for human dementia. Here, we will discuss the existing models, and what they have revealed about the genetic pathways and pathogenesis of different forms of dementia. We will also describe how to set up forward and reverse genetic screens in C. elegans, which can be used to identify additional genes and processes involved in dementia.
Key words
C. elegans genome-wide RNAiprotein aggregationα-synucleinLewy bodies1 Introduction
The free living nematode worm Caenorhabditis elegans (C. elegans) has been studied extensively as an experimental model organism by scientists from many fields of biological, agricultural, and medical research for over 40 years. The animal, introduced into molecular biology research by Sydney Brenner in 1963, lives in soil, where it feeds mostly on bacteria (1). It has a very short life cycle of about 3 days and a relatively short lifespan of about 2–3 weeks, which facilitates genetic dissection of the mechanisms that determine lifespan. Because of its anatomical and genetic simplicity, it has proven to be an excellent model organism. In fact, it is the most completely understood metazoan organism in terms of development, genetics, and nervous system (1–5).
The C. elegans nervous system represents the animal’s largest organ in cell numbers, being composed of 302 neurons. The morphology of the neurons is relatively simple and unbranched, most neurons having two or less processes. Owing to this simplicity, it has been possible to deduct their anatomical location and connectivity within the animal from electron microscopic images. This has enabled researchers to map the full circuitry of neuronal transmission in C. elegans, making it an unprecedented model for metazoan neurobiology. Although the C. elegans nervous system is fairly complex, it does not constitute an actual brain, merely a local concentration of neurons in the head region known as the nerve ring. Subtypes of neurons such as dopaminergic, GABAergic, and cholinergic neurons are present that are homologous to classes of neurons in vertebrates (6).
Reproduction of C. elegans is generally hermaphroditic by self-fertilization of eggs, which hatch outside of the animal. Larvae hatch from the eggs after all cell divisions are complete and go through four larval stages before entering the adult reproductive age after about 3 days. The soma consists of a constant 959 cells, of which each cell ends up in predetermined positions in the organs and tissues that make up the animal. The complex pattern of cell divisions and anatomical position is known as the C. elegans cell lineage (4,7). Much of what is currently known on the genetics of apoptosis has been elucidated in C. elegans by genetic studies of this cell lineage (7).
The basic mechanism of RNA interference (RNAi) was identified in C. elegans in 1998 by Andrew Fire and Craig Mello (8). Soon thereafter, the process of RNAi was extensively applied to silence expression of target genes, first in C. elegans, but also in other animal models and cells. The application of RNAi in C. elegans has greatly facilitated genetic research.
Initial RNAi experiments in C. elegans used injections of double-stranded RNA (dsRNA) in the gonad, leading to robust knockdown of mRNA levels in various cell types, thereby suggesting that RNAi was able to penetrate cell and tissue membranes. This indeed appeared to be the case when it was shown that RNAi can be obtained by soaking worms in dsRNA solution or feeding them on Escherichia coli (E. coli) expressing dsRNA (9–11). Feeding of E. coli expressing target gene dsRNA is now the most commonly used way of knocking down genes systemically in C. elegans. Such feeding has been used for rapid genome-wide RNAi screening, which has led to the discovery of comprehensive sets of genes involved in many genetic, cellular, and developmental processes, some related to human disease. The application of RNAi and its high-throughput capacity in genome-wide screens have revolutionized genetics research in C. elegans (10–13).
All the above has led to a thorough understanding of the animal’s behavior, genetics, molecular, cell, and neurobiology, and even molecular pathways that can increase and decrease the animal’s aging and lifespan. In addition, genetic mechanisms underlying human disease have been elucidated in C. elegans indicating its value as a model for human diseases, including age-related dementias. Here, we discuss the existing C. elegans models for dementia and provide examples of how these models can be used to screen for genes involved in the disease process, either by classical genetics or genome-wide RNAi.
2 Dementia
As described in earlier chapters, dementia is part of the pathologic spectrum of many sporadic as well as familial neurodegenerative diseases, such as Alzheimer’s disease (AD), frontotemporal lobar degeneration (FTLD), and dementia with Lewy bodies (DLB). C. elegans models for these diseases can be subdivided into at least two categories, including mutant strains to study endogenous pathways associated with disease and transgenic strains to study human disease-related genes. These models can be used for large-scale phenotypic, genetic, and drug screens to find genetic or chemical modifiers of pathways and phenotypes related to disease. These studies could, therefore, lead to a better understanding of disease pathology, the development of clinical diagnostic tools, and uncover therapeutic targets for treatment of dementia in humans.
In this chapter, we discuss the existing C. elegans models for dementias and large-scale screens that can be used to identify novel genetic and chemical components that modify disease phenotypes. We explain what these models have taught us about disease mechanisms, and what they have revealed from therapeutic and diagnostic perspectives.
2.1 C. elegans Counterparts of Human Dementia-Related Genes
Familial early-onset forms of AD are often caused by mutations in presenilin-1 and 2 (PSEN1 and PSEN2). These presenilin mutations are thought to change the ratio of the highly amyloidogenic 42-amino acid-long amyloid β (Aβ) peptide to the less toxic Aβ40 amino acid peptide variant of this disease-related protein (for review see (14)). In C. elegans, an ortholog for PSEN1 has been found in a screen for suppressors of the egg-laying defective phenotype in lin-12 (sel-12) gain-of-function worms (15). The sel-12 gene has a function similar to that in vertebrates, functioning mostly during embryonic development to facilitate Notch/lin-12 signaling. Apart from sel-12, the C. elegans genome encodes two other orthologs to human presenilins: a homolog of presenilin-1, hop-1, which is in fact more homologous to human PSEN2, and an ortholog spe-4, which has no obvious human counterpart (16). Sel-12 and hop-1 are highly similar to PSEN1 and PSEN2 in structure as well as in function (16,17).
Interestingly, genetic deletion mutants for sel-12 and hop-1 are viable, which allows for modifier screens to find components of the C. elegans PSEN1 and PSEN2 pathways. For example, mutations in multiple genes were found in forward mutagenesis screens for suppression of the sel-12/PSEN1 egg-laying phenotype (18–21). Two of them, spr-3 and spr-4, encode C(2)H(2) zinc-finger proteins that are similar to the human REST/NRSF (Re1 silencing transcription factor/neural-restrictive silencing factor) transcriptional repressors, involved in neuronal differentiation (18). Two others, spr-1 and spr-5, encode an ortholog of human CoREST, a co-repressor of REST, which can be substituted by human CoREST, and a homolog of p110b, which is another member of the CoREST co-repressor complex (19). In all, studies in C. elegans have provided important clues to the genetic context and regulation of presenilins.
In addition to these AD-related pathways, many other pathways have been investigated in C. elegans. For example, an ortholog of Aβ (apl-1), and at least three orthologs of genes encoded in loci associated with familial Parkinson’s disease (PD), PARK loci, are present and under investigation in C. elegans, including LRRK2/lrk-1, PINK1/pink-1, and Parkin/prk-1 (20–24).
2.2 Aggregation of Human Dementia-Associated Proteins
2.2.1 Amyloid β
In AD, Aβ forms amyloid fibrils, which accumulate in deposits known as plaques. To study aggregation of Aβ, the human Aβ peptide was overexpressed in C. elegans. By using the C. elegans Myosin/unc-54 muscle-specific promoter to drive expression, robust expression levels were obtained. A 189-base pair fragment was overexpressed coding for amino acids 1–42 of Aβ fused to a synthetic signal peptide driven by the unc-54 promoter. Transgenic worms expressing this transgene showed formation of aggregates with important characteristic of amyloid fibrils, as shown by Thioflavin S binding, an amyloid-specific dye, which has altered fluorescent properties on binding. In addition, paralysis of the nematodes, which can be easily distinguished by dissection microscopy, occurred indicating a specific toxicity of Aβ to the muscle cells (25). Interestingly, oligomeric species of Aβ were detected in these strains that might be similar to the neurotoxic Aβ-derived diffusible ligand (ADDL) (24).
The insulin/insulin growth factor (IGF)-1 like signaling (IIS) pathway strongly affects lifespan and stress resistance in C. elegans. Inhibition of daf-2/IGF-1 receptor by RNAi increases lifespan and stress resistance in nematodes. Mice deficient in the IGF-1 receptor also show increased lifespan, indicating the conservation of this mechanism (26). The effect of daf-2 partially depends on heat shock factor-1 (hsf-1), an important transcriptional regulator of heat shock genes. Interestingly, knockdown of the daf-2 gene in Aβ transgenic nematodes results in decreased paralysis, whereas hsf-1 knockdown increases paralysis, which is accompanied by alterations in the aggregation phenotype (27). It will be interesting to find out if these genetic pathways also affect Aβ aggregation and toxicity in mice and humans, which would directly link aging with the prevalence of disease related to Aβ aggregation.
2.2.2 α-Synuclein
Another protein that is associated with forms of dementia is the presynaptic protein α-synuclein. This protein provides a remarkable link between several neurodegenerative diseases, most of which involve dementia. It is the main constituent of protein inclusions in the brains of PD patients that represent the pathological hallmark of this disease. Such inclusions (known as Lewy bodies) also occur in a subtype of dementia called DLB, which is currently the second most prevalent cause of dementia. Importantly, DLB often has an overlap with Alzheimer’s pathology, and is sometimes difficult to distinguish from either AD or dementia related to PD.
Interestingly, α-synuclein mutations and multiplications of the α-synuclein locus have been found to cause familial forms of PD. One familial PD mutation, E46K, is also known to be associated with dementia (28). Some aspect of aggregation of α-synuclein is most likely directly involved in toxicity with the cells that degenerate in these diseases. The current hypothesis is that multimers (multiple associated molecules) of α-synuclein are somehow able to damage cells leading to degeneration. Although it is very likely that α-synuclein is directly involved in the pathogenesis, the mode of toxicity remains unknown. Physiologically, α-synuclein appears to regulate docking events of synaptic vesicles, likely by influencing the fusion of membranes (28–32).
To study expression and accumulation of a disease-related protein such as α-synuclein in a living animal, one can make the accumulation visible under a fluorescence microscope by fusion to a fluorescent molecule such as Yellow Fluorescent Protein (YFP). The C. elegans body wall muscle provides a system in which one can track accumulation of various disease-related aggregating proteins during aging (33,34). The muscle cells are easily seen under a microscope, and are amenable to RNAi by feeding (Fig. 1). In addition, the robust expression levels obtained by the muscle-specific unc-54 promoter yield high protein levels, which facilitate biochemical and cell biological analysis of protein expression.
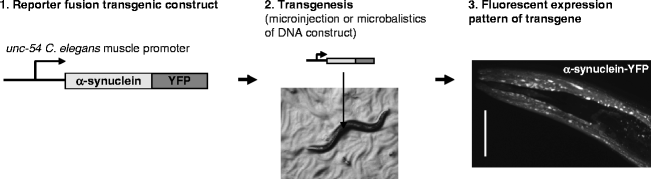

Fig. 1.
Tissue-specific transgene expression in transgenic C. elegans.
(1) A transgene is created using a known C. elegans tissue-specific promoter followed by the (human) gene of interest. By fusing the transgene to a fluorescent protein such as YFP (yellow fluorescent protein), its expression can be visualized. (2) Transgenic animals can be created by microinjection into the C. elegans gonad together with a marker or by microparticle bombardment. (3) Confocal laser scanning image showing head region of an adult C. elegans expressing α-synuclein fused to YFP in the body wall muscle using the unc-54 promoter element.
Transgenic strains expressing human α-synuclein fused to YFP in the body wall muscle show accumulation of the fusion protein during aging (35). Interestingly, the inclusions formed are mobile at young age, but show properties of immobilized aggregated protein at old age. To find genes involved in the formation of α-synuclein inclusion, a genome-wide screen was performed for genes that result in increased aggregation when knocked down by RNAi. Of the 80 genes found, genes that function in endomembrane-related compartments of the cell are overrepresented, which indicates a role for the endomembrane system in age-related synucleinopathies such as DLB (Fig. 2).
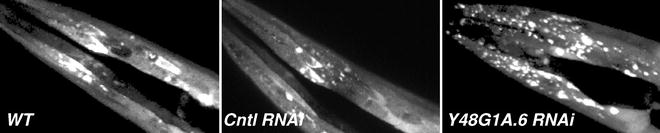

Fig. 2.
RNA interference of modifiers of α-synuclein inclusion.
Confocal images showing yellow fluorescent protein (YFP) fluorescence in the head region of a nematode expressing α-synuclein-YFP transgene in the body wall muscle. Worms are grown on regular food (wt, wild-type), HT115 bacteria containing empty RNAi vector (L4440) and bacteria containing Y48G1A.6 RNAi, to knock down one of the genes found in a genome-wide RNAi screen for modifiers of α-synuclein inclusion formation.
2.2.3 Tau (τ)
Tau (MAPT) is a microtubule-associated protein (MAP) mainly found in neuronal axons where it binds and stabilizes microtubules. In sporadic AD and familial frontotemporal dementia with Parkinsonism-17 (FTDP-17), a form of presenile dementia affecting the frontal and temporal cortex and some subcortical nuclei, hyperphosphorylated tau protein forms deposits in the brain. To model disease related to tau expression and deposition in C. elegans, Kraemer and colleagues have expressed both wild-type and mutated (301L and 337M) tau protein in C. elegans neurons (36). These nematodes developed a progressive phenotype of defective motility known as “uncoordinated phenotype” that can be distinguished by dissection microscopy, which was more apparent in the mutants. Interestingly, another characteristic of disease was captured in this model, since hyperphosphorylation of tau also appeared to occur in the transgenic lines (36). To identify modifiers of this toxicity, a genome-wide RNAi screen for enhancement of tau-related toxicity was performed (37). Some of the genes found are known to be involved in tau-related pathology, such as Glycogen synthase kinase 3β (GSK-3β) that can phosphorylate tau. New components have also been found, such as homologs of carboxyl terminus of Hsp 70-interacting protein (CHIP) and Heat shock cognate 70 (Hsc70) that cooperate in ubiquitination and proteasomal degradation. Thus, such genes might normally be involved in protection against tau-mediated toxicity.
Future work will show what modifier genes found in these genome-wide screens can tell us about the disease mechanism, and whether they represent diagnostic or even therapeutic targets to treat disease.
3 Genetics and Genomics
Although C. elegans are invertebrates, genetically they are similar to humans, and many disease-related genes and pathways found in humans have orthologs in C. elegans. These orthologous genetic pathways can be studied by making transgenics by microinjection or microparticle bombardment of transgenic constructs, or by conventional forward and reverse mutagenesis methods. For example, human disease-related genes or specific mutants, such as wild-type or mutant tau, Aβ, and α-synuclein, as described above, can be overexpressed in a tissue-specific manner using worm-specific plasmids created by Andrew Fire’s lab (Stanford University Medical Center, CA, USA) to study disease-related phenotypic effects on those cells.
< div class='tao-gold-member'>
Only gold members can continue reading. Log In or Register to continue
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


