Chapter 4
Molecular Infectious Disease Diagnostics
Serology or Culture Compared with Molecular Techniques
For immunologic testing, it is also important to consider whether the patient has had time to develop an immune response to a pathogen. This may take several weeks in reptiles.1 In acute infections, a specific immune response may not be present. There are large seasonal and temperature effects on the immune response of reptiles, as well as species differences.2–4 Other factors, including reproductive status and exogenous corticosteroid administration, may also affect development of a measurable immune response.5,6
When a clinician tests for an immune response, it is also important to consider what aspect of the immune system is being measured. A humoral immune response primarily produces antibodies to fight pathogens extracellularly, whereas a cellular immune response primarily uses cytotoxic T cells to trigger self-destruction of host cells infected with pathogens. Cellular and humoral immune responses are often found together but not always. An animal may have a cellular immune response without a significant humoral immune response and vice versa. Most common assays measure humoral immunity. A cellular immune response is typically most protective for intracellular pathogens such as viruses. Many intracellular pathogens elicit a relatively poor humoral immune response; a study of Tortoise herpesvirus 3 (TeHV3) in Greek Tortoises (Testudo graeca) found that only three of four experimentally infected animals developed detectable humoral immune responses, and the responses in the three that did show a reaction were relatively transient.7
Treating diverse herpesviruses as a single entity may lead to inappropriate management decisions. Before the diversity of tortoise herpesviruses was understood, a serosurvey of California Desert Tortoises using what was later shown to be TeHV3 as an antigen found that a number of California Desert Tortoises were seropositive; this was almost certainly cross-reactivity of antibodies to TeHV2 with TeHV3.8 From a population management perspective, it is crucial to know which viruses are endemic in a host species and the pathogenic implications of these viruses in other potential contact species.
Similarly, hemagglutination inhibition assays for ophidian paramyxoviruses in the genus Ferlavirus show poor concordance between different assays using different isolates.9 Recent work has expanded the known diversity of the genus Ferlavirus, and at least three distinct clades are known.10 The seroreactivity of samples from different snakes to different paramyxovirus strains differs significantly (Wellehan J, et al: unpublished data), and this may account, at least in part, for the lack of concordance seen. Overall, few pathogen clades exist whose diversity in reptiles is well enough understood that a clinician could be confident of the specificity of a detected antibody. This limits the current utility of serologic diagnostics in reptile medicine.
Culture of a pathogen from a diseased animal is considered the gold standard for identification of infectious agents whenever possible. For some bacterial pathogens such as coliforms and pseudomonads, isolation and identification is fairly straightforward. However, successful culture conditions have yet to be determined for the vast majority of microbes. Typically, around 10% of bacterial species can be cultured in a given ecologic niche,11 and success at culturing viruses or protozoa is even lower.
Even if an agent has been cultured, it is still necessary to identify it. Biochemical methods for bacterial identification have been the standard for the past century. These methods have been best developed for two phyla of bacteria: Proteobacteria, the “typical” gram-negative organisms, and Firmicutes, the “typical” gram-positive organisms. However, some of the most medically significant phyla in reptiles, such as Actinobacterium (containing Mycobacterium, Devriesea, Dermatophilus, and others) and Chlamydiae (containing Chlamydophila and others) are poorly identified by standard biochemical methods. Molecular methods involving nucleic acid sequencing are superior at identification.12 The contrast is even more stark in molecular versus classic fungal identification, in which identification has been morphologic, which misses significant cryptic diversity. The most significant fungal pathogens of reptiles are Chrysosporium sp. These are commonly called anamorphs of Nannizziopsis vriesii, but there is no support for this when sequence data from these isolates are examined. Sequence-based identifications of Chrysosporium sp. in reptiles include identification of a Chrysosporium sp. related to but distinct from Nannizziopsis vriesii in Bearded Dragons (Pogona vitticeps),13 Chrysosporium guarroi in green iguanas (Iguana iguana),14 and Chrysosporium ophiodiicola in a Black Rat Snake (Elaphe obsoleta).15 To date, there are no reports in which available sequence data support identification as Nannizziopsis vriesii. Sequence-based identification methods should be used to identify any agent where there is reason to question identification by conventional means.
Polymerase Chain Reaction
DNA is composed of two complementary strands oriented in opposite directions (Figure 4-1, A). Primers (short single-stranded segments of DNA) are designed to match the sequence of the two strands of pathogen DNA in a manner such that they face one another. The DNA is heated so that the two strands separate (Figure 4-1, B). The temperature is then decreased to an appropriate temperature for the primers to anneal (bind) with the DNA (Figure 4-1, C). If the primers do not match with the DNA sequence, they will not anneal and no product will be formed.
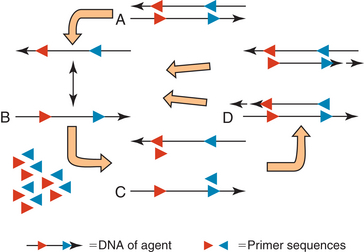
FIGURE 4-1 Polymerase chain reaction. A, Double stranded DNA of agent with known sequences (in red and blue) in opposite orientations on complementary strands. B, Nucleic acid is heated to separate the strands. C, DNA is cooled to an appropriate temperature for primers to bind to known sites. D, A high-temperature polymerase synthesizes a new complementary stand starting from the primer.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


