Fig. 1
Generic location of the six segments in the target DNA used to design LAMP primers. Forward (F3) and backward (B3) outer primers and forward (FIP) and backward (BIP) inner primers are indicated. The specially designed FIP and BIP primers contain two distinct sequences (F1c plus F2 and B1c plus B2, respectively) corresponding to sense and antisense segments of the target DNA, one for priming in the first stage and the other for self-priming in a subsequent amplification reaction stage
5.
Reagents for performing real-time PCR amplifications (e.g., SsoFast™ Evagreen® Supermix, Bio-Rad, CA, USA).
6.
Reagents for performing LAMP: Bst DNA polymerase large fragment and respective 10× reaction buffer (New England Biolabs), 25 mM of MgCl2, 20 mM of dNTPs, and 5 M of betaine.
7.
Reagents for performing gel electrophoresis: agarose, ethidium bromide, and 1× TBE buffer.
8.
Standard and real-time PCR equipments (e.g., CFX96™ Real-Time PCR Detection System, Bio-Rad, CA, USA).
9.
Standard equipment for performing agarose gel electrophoresis and the respective detection of the products under UV light.
3 Methods
3.1 Detection of T. annulata with Real-Time PCR
1.
Each amplification reaction is prepared for a final volume of 20 μl, including the addition of 5 μl of template DNA sample.
2.
Prepare a reaction mixture for all the DNA samples to test, including the positive and negative (water) amplification controls, containing 1× SsoFast™ Evagreen® Supermix, 0.3 μM of Tams1_forw primer and 0.15 μM of Tams1_rev primer, completing to 75 % of the final volume with PCR-grade water.
3.
Distribute 15 μl of the reaction mixture by individual 0.2 ml microtubes.
4.
Label each tube and add 5 μl of the correspondent DNA sample and controls.
5.
Proceed to the amplification step using the following program: (1) one initial denaturing step for 2 min at 95 °C and (2) 45 cycles of denaturation for 15 s at 95 °C, annealing for 30 s at 55 °C and extension for 30 s at 72 °C (see Note 8 ). The increase of fluorescence and amplification curves for each sample should be recorded and assessed according to the instructions of the manufacturer of the real-time PCR instrument. Only samples containing T. annulata DNA should yield positive amplification results.
6.
After the amplification step, an additional control step for the determination of the melting curve of the amplified fragments can be performed (if the real-time PCR instrument used has that feature). For this, at the end of the amplification program, add an additional step consisting of a 1 °C temperature increase every 5 s (beginning at 55 °C and ending at 95 °C) (see Note 9 ).
3.2 Detection of T. annulata with LAMP
1.
Each isothermal amplification reaction is prepared for a final volume of 15 μl, including the addition of 5 μl of template DNA sample.
2.
Prepare a reaction mixture for all the DNA samples to test, including the positive and negative (water) amplification controls, containing 4.8 U of the Bst DNA polymerase large fragment and 1× of the respective hybridization buffer, 6 mM of MgCl2, 1,400 μM of each dNTP, 0.8 M of betaine, 1.6 μM of each Tams1_FIP and Tams1_BIP primers, and 0.2 μM of each Tams1_F3 and Tams1_B3 primers, completing to 66.7 % of the final volume with PCR-grade water (see Note 10 ).
3.
Distribute 10 μl of the reaction mixture by individual 0.2 ml microtubes.
6.
Finalize the amplification step with an incubation at 80 °C during 2 min, to inactivate the enzyme.
7.
Confirm the occurrence of positive amplification results by analyzing the products in a 1.5 % (w/v) agarose gel electrophoresis (in 1× TBE buffer, at 90 V for 90 min; with ethidium bromide staining) and visualization under UV light, using standardized protocols (see Notes 13 – 16 ) (Fig. 2).
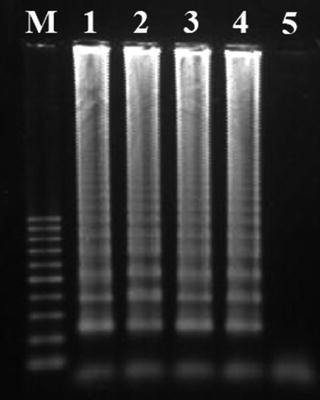

Fig. 2
Characteristic ladderlike appearance of LAMP products in an agarose gel electrophoresis. Lanes 1–4 correspond to positive amplification results and lane 5 corresponds to the non-template negative control. M—log scale 100 bp DNA Ladder (Jena Bioscience)
4 Notes
1.
Blood samples may be collected from animals into sterile tubes with EDTA and the total genomic DNA extracted using commercially available kits or automated systems (e.g., we use a BioSprint®96 automated workstation and the BioSprint®96 Blood kit, Qiagen) [9]. The average total DNA yield in samples is around 35–40 ng/μl. Extracted DNA should be stored at −20 °C until further use.
2.
As positive controls of amplification, we may use DNA samples extracted from T. annulata-infected macrophage cultures or directly from bovine blood samples for which T. annulata DNA was detected using other molecular tests such as reverse line blotting [9].
3.
Primers are frequently delivered lyophilized and need to be diluted with sterile water according to the manufacturer’s instructions. Stock solutions can be prepared at a standard concentration of 100 pmol/μl and stored at −20 °C. Aliquots of working solutions of each primer are prepared from stock solutions in water and stored also at −20 °C. Prepare small aliquots of working solutions (up to 100 μl) and avoid continuous freeze and thaw of the solutions.
4.




These primers were designed with complementary targets in conserved segments of the Tams1 gene, allowing the amplification of a fragment with about 319 bp. Alignments containing tens of Tams1 gene sequences were analyzed for detecting these conserved nucleotide segments within T. annulata, providing also enough nucleotide differences when compared to homologous genes from other closely related species [9]. The homologous genes of the closely related species T. lestoquardi, T. parva, and T. taurotragi all present several mismatches in the complementary target region for both primers, rendering it highly specific for T. annulata.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


