Cytogenetics and Infertility
Since the beginning of conventional chromosome analysis in the 1960s, the association between chromosomal abnormalities and reduced fertility has been observed in humans as well as in animals. This inspired the initiation of chromosome analysis in many domestic species. Research into animal cytogenetics reached its peak in the late 1980s when extensive studies of cattle, sheep, goat, river buffalo, pig, horse, dog, cat, and rabbit chromosomes were carried out in many laboratories worldwide.1,2 It was at this time that the first chromosome nomenclature systems for domestic animals were developed and the majority of chromosome aberrations described. Clinical cytogenetics received strong support from veterinarians, breeders, and owners because a direct connection between chromosome abnormalities and reduced fertility had been recognized. Even today, when whole genome sequences and high-throughput molecular tools are available for a number of domestic species, traditional cytogenetic screening of the breeding stock is still a part of evaluations.1,2
Compared with those of other domesticated species, the cytogenetics of alpacas, llamas, and camels has progressed slowly. Since the first description of the guanaco, Bactrian camel, and dromedary karyotypes 42 years ago, less than 20 reports in total have been published on normal and aberrant chromosomes in camelids.3 Furthermore, in other animal species, the inception of molecular cytogenetic methods (i.e., fluorescent in situ hybridization [FISH]) in the 1990s essentially improved the resolution and quality of chromosome analysis, but molecular cytogenetics in camelids is still in its infancy.4 The following discussion aims to give a comprehensive overview of the past and present knowledge about karyotyping and clinical cytogenetics in alpacas, llamas, and camels. The reasons that have hindered chromosome studies in these species will be analyzed and solutions for improvement proposed.
Alpaca, Llama, and Camel Chromosomes
The Species
The family Camelidae, which belongs to the mammalian order Cetartiodactyla, was separated from other even-toed ungulates about 40 to 45 million years ago.5,6 Thereafter, as revealed by mitochondrial deoxyribonucleic acid (DNA) analysis, the family was split into the genera Camelus and Lama, which occurred in North America about 11 million years ago.5 Today, two Camelus species remain in Asia and North Africa: (1) the two-humped Bactrian camel (Camelus bactrianus [CBA]) and (2) the one-humped dromedary camel (Camelus dromedaries [CDR]). The Lama genus is represented by four South American species: the domesticated alpaca (Lama pacos [LPA]) with its possible wild ancestor the vicugna (Lama vicugna [LVI]) and the domesticated llama (Lama glama [LGA]) with the probable wild ancestor the guanaco (Lama guanacoe [LGU]). Compared with other domesticated species, the evolutionary history of the Camelus and Lama genera remains elusive. According to archeologic dating, the earliest known two-humped camel domestication occurred in China and Mongolia 5000 to 6000 years ago, and llamas and alpacas were domesticated in the high Andes around 9000 years ago.7,8 Thus, the camelids have been associated with humans for almost as long as cattle, horses, and dogs.
Chromosome Number and Karyotypes
Although preliminary studies to visualize the chromosomes and assess the modal number in cattle, horses, rabbits, and pigs were carried out in the early 1900s, the first reports about the karyotypes of camelids emerged only in the 1960s.9 This was the time when essential methodologic advances such as improved cell culture methodology and the invention of hypotonic treatment in human cytogenetics inspired and promoted cytogenetic programs in many species, including the alpacas, llamas, and camels.10,11
The first two reports proposed the alpaca diploid chromosome number to be 2n = 72, which was soon corrected to 2n = 74.3,12–16 It also became apparent that extensive similarities exist in the chromosome size and morphology between all extant camelid species, that is, the vicugna and the Bactrian camel, and the guanaco, the dromedary, and the Bactrian camel.3,14–16 These early results were confirmed by all of the subsequent studies leading to the overall conclusion that despite distinct anatomic and physiologic differences and very specialized adaptations, karyotypes of the New World and Old World camelids have remained extremely conserved in the course of evolution.17–19
Chromosome Measurements
To date, only two attempts have been made to measure the various parameters of alpaca chromosomes, such as relative length, arm ratio, and centromeric index.20,21 The results of these two studies differ from each other and do not give a clear overview about the sizes and morphologic categories of the chromosomes. For example, in one study, the largest autosome makes up 4.2% of the genome, and in another, 5.9%.20,21 The authors, however, agree about the relative sizes of the smallest autosomes, which are 0.9% and 0.8%, respectively. Discrepancies with regard to the sex chromosomes also exist. Zhang and colleagues (2005) showed that the X chromosome is metacentric (centromeric index 48%), with a relative length of 3.6.20 Measurements carried out by Di Berardino and colleagues (2006) classified the X chromosome as submetacentric (centromeric index 40%), comprising 4.9% of the genome.21 The observed differences might have occurred because in one study, metaphase was analyzed and in another prometaphase chromosomes were analyzed.20,21 It is, however, more likely that these discrepancies can be attributed to the inherent properties of the camelid karyotype. As the chromosomes are composed of a high number, gradually decrease in size, and are mostly biarmed elements (Figure 21-1), it is difficult to delimit one morphologic type of chromosome from another.21

Figure 21-1 Routinely Giemsa-stained metaphase spread (left) and a corresponding karyotype (right) from a normal male 74,XY (A) and a normal female 74,XX (B) alpaca. Chromosomes are arranged arbitrarily following the nomenclature by Balmus et al. (Balmus G, et al: Cross-species chromosome painting among camel, cattle, pig and human: further insights into the putative Cetartiodactyla ancestral karyotype, Chromosome Res 15:499-515, 2007; Our unpublished data.)
Banding Techniques Applied to Study Camelid Chromosomes
In all of the early studies, the chromosomes were visualized by routine staining with Giemsa.3,14–16 This allowed counting of the chromosomes and rough arrangement into a karyotype based on size and morphology. Even after the development of chromosome banding techniques, routine staining with Giemsa has remained a useful method for quick karyotyping in clinical cytogenetics (see Figure 21-1) and for carrying out chromosome measurements.20,21
GTG-Banding
GTG-banding (G-banding) by Trypsin and Giemsa staining is a procedure in which chromosomes are first treated with trypsin to differentially remove chromatin proteins and then stained with Giemsa.22 This method, which produces a distinct banding pattern for each chromosome pair, was first used in the Bactrian camel, the guanaco, and the llama, followed by a detailed G-banding comparison of all four South American camelid (SAC) species.17,18 More recently, high-resolution GTG-banded karyotypes have been published for the alpaca and the dromedary.20,21,23 All studies agree about one point: Besides sharing the same chromosome number and morphology, all camelids have also very similar or identical G-banding patterns.
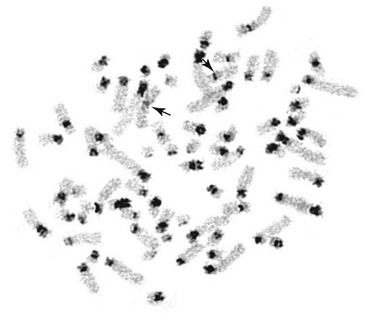
C-Banding
C-banding is a method in which chromosomes are subjected to acidic and alkaline hydrolysis, treatment with hot salt, and Giemsa staining.24 These treatments remove most of the chromatin, leaving constitutive heterochromatin intact for staining only (Figure 21-2). All camelid chromosomes show C-positive bands not only at the centromeres but also on the short arms of acrocentric chromosomes.17,18,21 Although the C-banding pattern across different camelid species is the same, considerable C-band polymorphism has been observed among chromosome pairs as well as between homologs.21 These observations suggest that the variable size of constitutive heterochromatin could explain the discrepancies between different reports in determining arm ratio and classifying the chromosomes.21 However, because of variations in the size and intensity of C-bands, the method is not useful for routine identification of autosomes or sex chromosomes.
Nucleolus-Organizing Regions
The nucleolus-organizing regions (NORs) are chromosomal sites of 5.8S, 18S, and 28S ribosomal ribonucleic acid (RNA; ribosomal RNA [rRNA]) genes and were originally detected by silver-staining (Ag-I) technique or NOR-banding.25 The method uses ammoniac silver nitrate solution and stains the chromosomal sites containing actively transcribed rRNA genes. The NORs may also be studied by using radioactive or fluorescence in situ hybridization (ISH; or FISH) with rDNA probes, which detect all rRNA loci regardless of their transcriptional status.
In camelids, the NORs are located on the short arms or secondary constrictions of four, five or six chromosome pairs.17,18,21,26,27 No differences in the number of Ag-positive NORs and those detected by radioactive in situ hybridization with 18S DNA were observed.26 However, as expected, the nomenclature of NOR chromosomes is different in all five studies, which further underlines the challenges in identifying camelid chromosomes. Last but not least, localization of NORs also represents the first gene mapping results in these species.
Other Banding Methods
Besides routine staining with Giemsa, GTG-banding, C-banding, and NOR-banding, the use of other differential banding techniques in camelid cytogenetics has been limited. Comparison of GTG-banding, RBG-banding, and RBA-banding patterns in alpaca chromosomes has been carried out in only one study. 21The RBG-banding and RBA-banding methods produce a reverse banding pattern compared with GTG-banding, whereas in the RBG-banding method, the chromosomes are stained with Giemsa and in the RBA-banding method, the chromosomes are stained with acridine orange. The study reports a good correspondence between the G-banding and R-banding patterns in the alpaca. Additionally, to better analyze the observed polymorphism of constitutive heterochromatin (see above), sequential QFQ-banding or C-banding was carried out in the alpaca.21 The authors concluded that because of the significant variation in the amount of heterochromatin, pronounced variation exists in the size of the short arms of many chromosomes, making conventional karyotyping in alpacas and other camelids unreliable and ambiguous.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree