CHAPTER 43 The Feline Genome and Clinical Implications
Cytogenetics
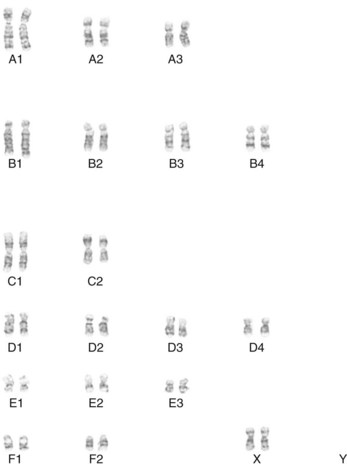
Early studies of mitotic chromosomes of the domestic cat revealed an easily distinguishable karyotype consisting of 18 autosomal chromosomes and the XY sex chromosome pair, resulting in a 2N complement of 38 chromosomes for the cat genome (Figure 43-1).57–60 Cat chromosomes are fortuitously easily distinguishable, clearly defined by size, centromere position, distinctive giemsa banding patterns of the short (p) and long (q) arms of each chromosome, and the presence of only a few small acrocentric chromosomes, which have no p arms and are traditionally hard to distinguish. Various cytogenetic techniques, such as R-, RBG-banding and fragile site studies, have also helped distinguish and characterize the cat chromosomes.47–4952 For example, cats do not have a significant fragile X site on the X chromosome that is found in humans and is associated with mental retardation. Although a sequential numbering of the chromosomes has been suggested,3 the historical classification of chromosomes into morphologic groups has been retained in the cat. Hence cats have three large metacentric chromosomes (A1 to A3), four large subtelomeric chromosomes (B1 to B4), two medium-size metacentrics (C1 and C2), four small subtelomerics (D1 to D4), three small metacentrics (E1 to E3), and two small acrocentrics (F1 and F2). The X chromosome is midsize and subtelomeric, similar to chromosome B4.
Early chromosome staining recognized some major alterations in the felid genome, particularly the Robertsonian translocation of F1 and F2 to form chromosome C3 in the ocelot lineage of cats from South America (2N = 36).59 Minor pericentric inversions, additions, or deletions of the small chromosomes cause variation in the felid karyotype. The pericentric inversion of chromosome F1 produces a small, more centromeric chromosome and represents as E4 in many cat species. Overall, domestic cats have a chromosomal architecture that is highly representative for all felids and ancestral for most carnivores.34,44
Historically, the first genetic consideration to explain reduced fertility or intersex cats is chromosomal differences, especially the loss of one of the sex chromosomes. Karyotypic and now gene-based assays are common methods to determine if a cat with ambiguous genitalia50 or a poor reproductive history has a chromosomal abnormality. Karyotypic studies of male tortoiseshell cats have shown that they are often mosaics, or chimeras, being XX/XY in all or some tissues.* The minor chromosomal differences that are cytogenetically detectable between a domestic cat and an Asian leopard cat are likely the cause of fertility problems in the Bengal cat breed, which is a hybrid between these two species. Other significant chromosomal abnormalities causing common “syndromes” are not well documented in the cat.
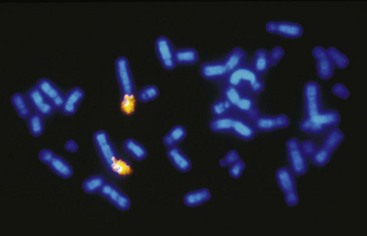
The sufficient variation of cat chromosomal sizes also allowed for the easy flow sorting of cat chromosomes.56 The DNA in the flow-sorted pools of each chromosome could be individually dye labeled. The dye-labeled DNA from each chromosome could then be hybridized to mitotic chromosomes of another species, such as humans, which provided a gross overall view of which chromosomes between the two species had the same DNA (Figure 43-2). For example, the p arm of human chromosome 1 (1p) is largely composed of the same genes that are on the cat chromosome defined as C1, whereas human chromosome 1q is composed of genes that are found on cat chromosome F1. The chromosome painting technique could also be performed reciprocally, implying painting cat chromosomes onto human mitotic chromosome spreads and human chromosomes onto cat mitotic chromosome spreads, revealing the high conservation of chromosomal arrangement of cat to humans,56,61 specifically compared to mice.53 Thus chromosome painting gave an excellent overview of cat genome organization,38 which greatly facilitates candidate gene approaches because the location of particular genes could be anticipated in cats from comparison with the genetic map of humans.55 This additional confirmation of conservation to human, with regard to genome organization, further supported additional genetic resource development for the cat as a valuable animal model for human disease.
Genetic Maps
Somatic Cell Hybrid
The aesthetically pleasing karyotype supported the early somatic cell hybrid genetic maps of the cat.35,37 A somatic cell hybrid is a fusion of the cell lines, generally fibroblasts, of two different species. The cell line of one species—usually a rodent, such as a mouse or Chinese hamster—is compromised in some way, such as by having an enzyme deficiency that causes growth incompatibility in non-supplemented media. The cell line from the species of interest, in this case the cat, is damaged by a different, chemical means. The fusion of the two compromised cell lines leads to the chromosomes of the cat integrating into the nucleus and sometimes the chromosomes of the rodent cell line, which then rescues the rodent cell line as a functional enzyme is now present to support the growth of fusion cells. Many different fusion lines are maintained and propagated; however, the entire complement of cat chromosomes is never completely retained in a given cell line. Thus a given cell line will have all the rodent chromosomes, which happen to be mainly acrocentric chromosomes, and only one or a few cat chromosomes. A proper somatic cell hybrid panel would have a representation of at least each of the cat chromosomes in the set of fusion cell lines. Analysis of the mitotic chromosomes of the cell lines can often show which specific cat chromosome may be present because the cat chromosomes are clearly distinguished from those of rodents by size and shape. These cell lines can then be assayed for the presence or absence of specific proteins, or DNA sequences, thereby indicating that the genes that create the proteins or are represented by the DNA segments must reside on the cat chromosome that is within the cell line. This mapping approach provided the first rudimentary genetic map of the cat with 105 different loci,35 including the association for the genes for hemoglobin beta (HBB) and tyrosinase (TYR).36 The HBB polymorphism was shown to be associated with the Siamese coloration, also known as points. Recently, this coloration has been proved to be a mutation in TYR.13,23,51
Recombination Map
Interspecies Hybrids
During the late 1960s and early 1970s, the role of viruses in cancer etiologies was under intense investigation, and the cat figured largely in these studies. Feline leukemia had been shown to be caused by a virus (FeLV); hence the cat became an important model for viral carcinogenesis. Because leopard cats, a small and rather abundant type of wildcat from Asia, were shown to be resistant to FeLV infection,43 genetic studies relating to viral carcinogenesis initiated with domestic and Asian leopard cats.1 Although viral carcinogenesis did not play as significant a role in cancer etiology as was initially anticipated, the role of the leopard cat in cat genetics and genomics was crucial.
Genetic recombination–based maps of the cat are an improvement in resolution over somatic cell hybrid maps, with the added benefits of estimating gene order and distance between genes on a chromosome, not just presence or absence. The Bengal cat breed has been influential in the construction of the first genetic maps of the cat. Bengals are a hybrid between domestic cats, primarily Abyssinians and Egyptian or Indian Maus, and a different species of felid, the Asian leopard cat (Felis [Prionailurus] bengalensis). The breed was developed in the late 1960s15 and is now one of the most popular breeds in the world, although not all registries recognize it. The evolutionary distance between the parental type cats of the Bengal breed is significant.16,17 The millions of years of evolution between a leopard cat and a domestic cat have made the DNA sequence of each gene more genetically diverse than the gene sequence found between any two domestic cats or any two leopard cats. Thus a pedigree consisting of the first-generation Bengals (F1) and a second-generation backcross to one of the parental-type cats was the basis of the first recombination map for the cat.26 Genetic variation is required to build a genetic map based on recombination, and because these Bengal crosses would generate offspring with very high genetic polymorphism, the interspecies cross was efficient. The first version of feline interspecies hybrid-based linkage map contained approximately 250 microsatellite markers (also known as short tandem repeats [STRs]).26 This map was effective for the initiation of pedigree studies for families segregating for particular phenotypic traits or diseases. Although rudimentary, the interspecies backcross map assisted targeted candidate gene approaches—in particular, the discovery of the mutation that causes feline polycystic kidney disease (PKD).62 The genetic map also led to the discovery that a chromosomal rearrangement involving the gene LIX1 causes spinal muscular atrophy in the Maine Coon cat.8,12
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree