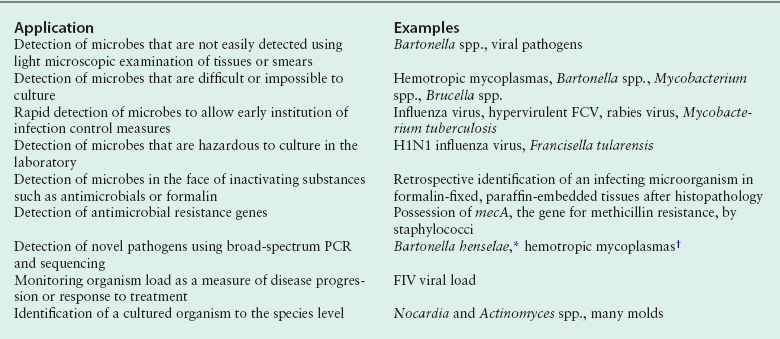
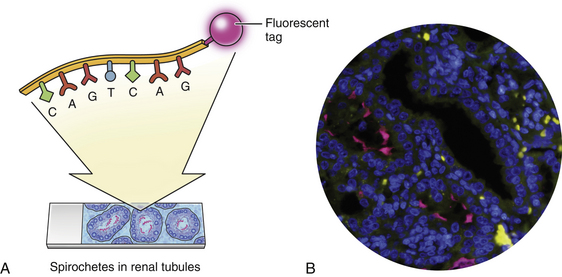
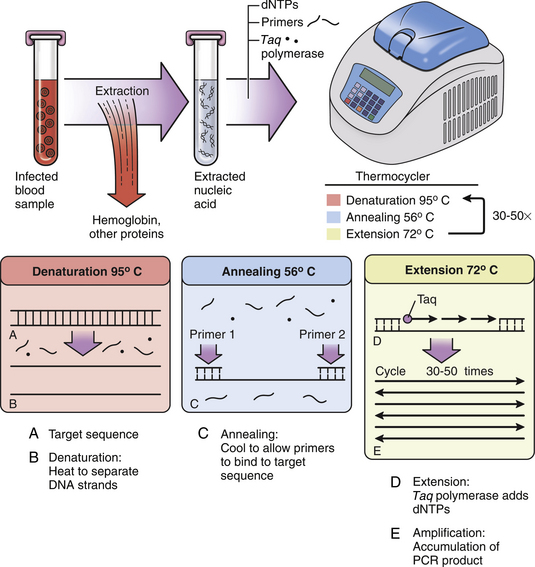
Chapter 5 • Assays for detection of nucleic acid such as polymerase chain reaction assays are increasingly available commercially for diagnosis of companion animal infectious diseases. • Molecular diagnostic assays include those that involve amplification (the copying of nucleic acid so that it can be more readily detected) or probe-based assays that do not involve nucleic acid amplification. • Specimens for these assays should be collected using sterile technique in order to minimize contamination. • Interpretation of the results of molecular diagnostic tests requires knowledge of the problems and pitfalls of each type of assay. The results of testing at one laboratory may differ from that at another because of variations in assay design. • With increased automation, laboratory proficiency testing, improved quality assurance, and standardization of reporting, the availability and utility of molecular diagnostic tests for companion animal infectious diseases will increase. DNA or RNA probes are short, single-stranded nucleic acids (usually <50 base pairs) that have a sequence that hybridizes to (i.e., is complementary to) a specific segment of DNA from the target microorganism (Figure 5-1). For in situ hybridization, the probe is labeled with a fluorescent or chemical tag so that the bound probe can be detected. The probe is reacted with a tissue specimen on a microscope slide in order to determine if the organism is present in the specimen. For PCR, two specific short (approximately 20 bases) DNA sequences called primers, which are complementary to the target microorganism’s DNA, are used in conjunction with a machine called a thermocycler to amplify a specific segment of DNA from just one copy to millions of copies that can be more easily detected. In situ PCR is a combination of these two techniques.1 FIGURE 5-1 Fluorescence in situ hybridization (FISH). A, A nucleic acid probe that is complementary to a segment of leptospiral DNA is tagged with a fluorescent label and incubated with a tissue section on a slide. B, Unbound probe is washed away, and the section is examined using fluorescence microscopy. (Leptospira FISH image courtesy Dr. Richard Goldstein.) Nucleic acid detection methods are ideally suited to detection of organisms that are not easily found using cytology or histopathology, are slow-growing, or are difficult to culture, as well as when a rapid (<12 hours) diagnosis is required. Other applications are shown in Box 5-1. The use of nucleic acid–based assays for diagnosis has increased dramatically in veterinary medicine over the past 10 years. Because of the exquisite sensitivity of some assays, especially those that involve DNA amplification, positive results may reflect contamination that occurs in the laboratory. Contamination problems have decreased with increased automation and the use of real-time PCR assays (see later). In human medicine, proficiency testing programs have helped to overcome quality assurance problems in laboratories that perform molecular diagnostic assays.2 Not all PCR assays for a specific pathogen are equal, because the nucleic acid primers and probes and the equipment used to run each assay may be different. So a PCR assay for Leptospira performed in one laboratory may perform differently than an assay used in another laboratory. A plethora of nucleic acid detection methods have been developed since the advent of PCR, many of which are commercially available for diagnosis of human infectious diseases but are not yet being widely used in veterinary medicine. These are described in detail elsewhere.1,3,4 The purpose of this chapter is to outline (1) the indications for nucleic acid detection methods, (2) the best ways to collect and transport specimens for these assays, (3) the most commonly used methods in veterinary medicine, and (4) guidelines for their interpretation. The recommendations for specimen collection and transport presented here are based on the guidelines published by the Clinical and Laboratory Standards Institute (CLSI).5 These guidelines are updated on a regular basis and ensure that detection of nucleic acids within clinical specimens is optimal. In some instances, nucleic acid may be present and detectable in a specimen even when the specimen has not been handled in a manner that is consistent with the guidelines. Many laboratories publish their own specific guidelines for specimen collection and transport. Specimens should be properly packaged and labeled (see Chapter 3). In situ hybridization is generally performed on formalin-fixed, paraffin-embedded tissue specimens. Specimens that have been archived for several years may still be adequate, even for detection of RNA, which is more labile than DNA.6,7 The optimum specimen for fluorescence in situ hybridization (see later for discussion) is tissue that has been snap frozen in liquid nitrogen after being embedded in optimal cutting temperature (OCT) medium.5 When collecting specimens for PCR, the timing of collection relative to the course of disease and the best specimen type must be considered. For acute diseases, PCR is often most useful early in the course of disease. For chronic diseases, timing is less critical. Knowledge of organism shedding patterns can help to determine the most appropriate timing of specimen collection. For example, Leptospira organisms are primarily present in blood in the first week of acute illness, after which they may be shed in the urine.8 In general, for detection of pathogens that contain DNA (i.e., all bacteria, fungi, some viruses), either fresh or frozen specimens should be submitted. In general, DNA is stable in tissue for up to 24 hours at 2°C to 8°C, at least 2 weeks at −20°C, and at least 2 years at −20°C or below −70°C.5 Specimens held at room temperature should be submitted immediately and reach the laboratory within 24 hours. Specimens can also be refrigerated and submitted on wet ice within 72 hours. Provided the target of the assay is not an erythrocyte pathogen, erythrocytes should be removed before storage if possible. Specimens should not be stored in frost-free freezers, as these undergo repeated freeze-thaw cycling, which can be associated with DNA degradation.9 Box 5-2 outlines recommendations based on specimen type. For tissue specimens, the optimal amount is 1 to 2 g, although more or less may be required depending on the cellularity of the specimen.5 Tissues stored in formalin, especially for prolonged periods, are suboptimal for PCR because the formalin cross-links the DNA in the specimen. Tissues fixed briefly in formalin and then paraffin-embedded are preferable to tissues stored in formalin, because the formalin is removed during the embedding process. Paraffin-embedded specimens should only be used when no other specimens are available.5 Although PCR can be performed on feces, its sensitivity may be lower than with other specimens because inhibitors of PCR are particularly abundant in fecal material.10 The use of appropriate amplification controls by the laboratory facilitates detection of false negatives due to the presence of inhibitors. Probe hybridization can be performed on a nitrocellulose membrane (solid-phase hybridization); on formalin-fixed, paraffin-embedded sections mounted on a microscope slide (in situ hybridization); or in solution (liquid-phase hybridization). For solid-phase hybridization, the probe is reacted with microorganism DNA that has been immobilized on the membrane. Unbound probe is washed away, and the bound probe is detected using fluorescence, chemiluminescence, radioactivity, or color development (in the same way that bound antibody or antigen is detected in an ELISA or immunofluorescent antibody assay). For in situ hybridization, formalin-fixed, paraffin-embedded specimens are sectioned and mounted on a special slide. The sections are deparaffinized, dried, and incubated with a solution that contains the probe, so both the presence and the location of the target pathogen within tissues can be identified. In situ hybridization assays that include a fluorescent-labeled probe are referred to as fluorescence in situ hybridization (FISH) assays (see Figure 5-1). Because liquid-phase hybridization occurs in solution, unbound probe cannot be washed away. To overcome this problem, a chemiluminescent acridinium ester label is attached to the probe. A subsequent chemical hydrolysis step selectively degrades only unbound probe. On addition of peroxides, the intact (hybridized) probe then emits light.11 Although not yet widely used for veterinary applications, peptide nucleic acid (PNA) probes are now increasingly available to detect target DNA. PNA probes are uncharged peptides that mimic DNA and bind to complementary DNA sequences just as a nucleic acid probe would.12,13 PNA probes lack the net negative charge of nucleic acid probes; therefore, the electrostatic repulsion that normally occurs when two negatively charged DNA strands hybridize does not occur. The result is a more stable and specific binding of the probe to its target, which in turn can be associated with increased assay sensitivity and specificity. Branched DNA assays and hybrid capture assays are highly sensitive hybridization methods that include steps to intensify the signal generated from probe hybridization.3,4 They are not yet widely used in veterinary medicine. PCR allows the specific amplification of DNA sequences from just one copy to millions of copies, which can be more readily detected. The DNA from a clinical specimen is extracted using a commercially available DNA extraction kit. A pair of primers, roughly 20 nucleotides long, is then used to bracket a desired DNA sequence, which is subsequently copied using a DNA polymerase enzyme (Figure 5-2). FIGURE 5-2 Basic PCR assay. Nucleic acid is extracted from a clinical specimen through removal of contaminating proteins such as hemoglobin. Two primers are added that hybridize at each end of a specific segment of a pathogen’s DNA. Also present in the reaction mixture are the enzyme Taq polymerase (dots) and free nucleotides (dNTPs) for new DNA. A thermocycler is used to rapidly and repeatedly heat and cool the tubes through denaturation, annealing, and extension steps. Denaturation separates double-stranded DNA. Annealing occurs at a specific temperature that causes the primers to bind only to their target sequences. When the reaction mixture is then heated to 72°C, the Taq enzyme uses the primers as initiation points for DNA extension, and the target sequence is copied. The process is repeated 30 to 50 times, with logarithmic accumulation of the PCR product, so there are millions of copies of the target pathogen’s DNA. These can then be detected using agarose gel electrophoresis (see Figure 5-3).
Nucleic Acid Detection Assays
Introduction
Specimen Collection and Transport
Nucleic Acid Probe Assays
Polymerase Chain Reaction
Diagnostic Methods
The Polymerase Chain Reaction
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Nucleic Acid Detection Assays
Only gold members can continue reading. Log In or Register to continue