Primer/probe
Sequence (5′–3′)
Complementary target
F_Actin
GGC TCY ATY CTG GCC TC
β–actin gene of mammalsa
R_Actin
GCA YTT GCG GTG SAC RAT G
P_Actinb
Cy5.5-TAC TCC TGC TTG CTG ATC CAC ATC-BHQ2
F_IS6110
GGG TCG CTT CCA CGA TG
IS6110 element of MTC speciesc
FN_IS6110
CTC GTC CAG CGC CGC TTC GG
R_IS6110
GGG TCC AGA TGG CTT GC
P_IS6110 d
FAM-CGC GTC GAG GAC CAT GGA GGT-BHQ1
F_16SrDNA
CCG CAA GGC TAA AAC TCA AA
Mycobacterial 16S rDNAe
R_16SrDNA
TGC ACA CAG GCC ACA AGG GA
P_16SrDNAf
TET-TCG ATG CAA CGC GAA GAA CCT TAC-BHQ1
F-esat6
AGG CGT ACC AGG GTG TC
RD1 (Rv3875 locus, esat6 gene)
R-esat6
CGA AGC CAT TGC CTG ACC
P-esat6 b
Cy5.5-ACAACGCGCTGCAGAACC TGG-BHQ2
F-Rv1510
CCT GCA AGA AAC GAC CCG
RD4 (Rv1510 locus)
R-Rv1510
GCGACGGTGCCAATCATC
P-Rv1510f
TET-CCATCGTACCCATCCGCT GCG-BHQ1
F-Rv2073c
AGTCGGTGTGCACGATGG
RD9 (Rv2073c locus)
R-Rv2073c
CGC TCG TTG CCG AGC AC
P-Rv2073cg
Texas Red-CTG GTC GCC GAG TAT CCC GAA G-BHQ2
7.
Reagents for standard PCR amplification: Taq DNA polymerase and respective 10× reaction buffer; deoxynucleotide triphosphates (dNTPs); MgCl2.
8.
Reagents for real-time PCR amplification (e.g., the SsoFast supermix, BioRad).
9.
Homogenizer/bead shaker (e.g., the FastPrep FP120 Bio101, Savant Instruments Inc., Holbrook, NY).
10.
Centrifuge.
11.
Standard and real-time PCR equipment (e.g., the CFX96 real-time PCR instrument, BioRad).
3 Methods
To prevent the risk of human infection, the manipulation of MTC cultures and TB-suspect tissues must be performed in a confined biosafety level 3 laboratory.
3.1 DNA Extraction from Reference and Clinical MTC Cultures
The culture supernatants can be processed using a rapid and simple boiling-based DNA extraction procedure.
1.
Grow the MTC strains using standard liquid culture media and procedures for these organisms.
2.
After incubation, centrifuge 10 ml of culture at 3,800 × g for 30 min.
3.
Discard the supernatant, wash the pellet in 10 ml of PBS, and centrifuge again at 3,800 × g for 30 min.
4.
Discard the supernatant and suspend the pellet in 250 μl of TE buffer.
5.
Heat the suspension in a water bath at 95 °C for 25 min.
6.
Centrifuge at 15,000 × g for 5 min.
7.
Transfer a 150 μl aliquot of the supernatant (containing the DNA) to a sterile microtube and store at −20 °C until further use.
8.
Stock DNA suspensions are diluted ten times in distilled water before its use as template for PCR assays.
3.2 DNA Extraction from Animal Tissue Samples
1.
Homogenize the tissue sample using a clean, sterilized pestle and mortar (the addition of a small volume of PBS buffer and sterilized powdered glass, silica, or sand can help the homogenization process).
2.
Transfer 450 μl of tissue suspension to screw-cap microcentrifuge tubes.
3.
Inactivate the tissue sample in a water bath at 100 °C for 5 min.
4.
Centrifuge the samples in a bench centrifuge at 15,000 × g for 2 min.
5.
Discard the supernatant and add 80 μl of PBS buffer and an equivalent volume of 100 μl of zirconium beads.
6.
Proceed to the mechanical disruption of the mycobacterial cells (e.g., FastPrep FP120 Bio101 bead shaker at 6.5 m/s for 45 s, three times).
7.
Cool the suspensions on ice for 15 min.
8.
Proceed to the DNA extraction using a commercially available kit, according to the manufacturer’s instructions (e.g., the tissue protocol of the QIAamp DNA Mini Kit).
9.
Store the genomic DNA suspensions at −20 °C until further use.
10.
Stock DNA suspensions are diluted ten times in distilled sterile water before its use as template for PCR assays.
3.3 Detection of MTC in Tissue Samples by Nested Real-Time PCR
The nested IS6110-targeted real-time PCR assay consists of two steps: (1) a first standard PCR using primers FN_IS6110 and R_IS6110 and (2) a second duplex real-time PCR using the previous amplification product as template and a mixture of IS6110 and β–actin gene-targeted TaqMan ® probes (P_IS6110 and P_Actin, respectively) and the corresponding flanking primers (F_IS6110/R_IS6110 and F_Actin/R_Actin, respectively) (Table 1) (see Notes 2 and 3 ).
1.
For the first standard PCR, each amplification reaction is prepared for a final volume of 25 μl, including the addition of 5 μl of template DNA sample.
2.
Prepare a reaction mixture for all the DNA samples to test, including the positive (DNA from a reference strain culture) and negative (distilled sterile water) amplification controls, containing 400 μM of dNTPs, 1 U of Taq DNA polymerase and 1× of the respective buffer, 3.5 mM of MgCl2, and 0.8 μM of each primer (FN_IS6110 and R_IS6110), completing to 80 % of the final volume with PCR-grade water (see Note 4 ).
3.
Distribute 20 μl of the reaction mixture by individual 0.2 ml microtubes.
4.
Label each tube and add 5 μl of the correspondent DNA sample and controls.
5.
Proceed to the amplification step using the following program: (a) initial denaturation step at 95 °C for 10 min; (b) 45 cycles of 30 s at 95 °C, 30 s at 65 °C, 30 s at 72 °C; and (c) a final extension step of 10 min at 72 °C.
6.
7.
For the second nested duplex real-time PCR, each amplification reaction is prepared for a final volume of 20 μl, including the addition of 5 μl of the previous amplification product (including for the correspondent positive and negative amplification controls).
8.
Prepare a reaction mixture for all the DNA samples to test containing: 1× SsoFast supermix, 0.4 μM of each primer (F_IS6110, R_IS6110, F_Actin, and R_Actin), and 0.15 μM of each TaqMan ® probe (P_IS6110 and P_Actin), completing to 75 % of the final volume with PCR-grade water.
9.
Distribute 15 μl of the reaction mixture by individual 0.2 ml microtubes.
10.
Label each tube and add 5 μl of the correspondent DNA sample (product of step 6) and controls.
11.
Get Clinical Tree app for offline access

Proceed to the amplification step using the following program: 1 cycle at 95 °C for 2 min, followed by 45 cycles at 95 °C for 5 s, and 60 °C for 10 s. The increase of fluorescence and amplification curves for each sample should be recorded and assessed according to the instructions of the manufacturer of the real-time PCR instrument. Only samples containing MTC DNA should yield positive amplification results with the IS6110-targeted probe (see Note 2 and Fig. 1).
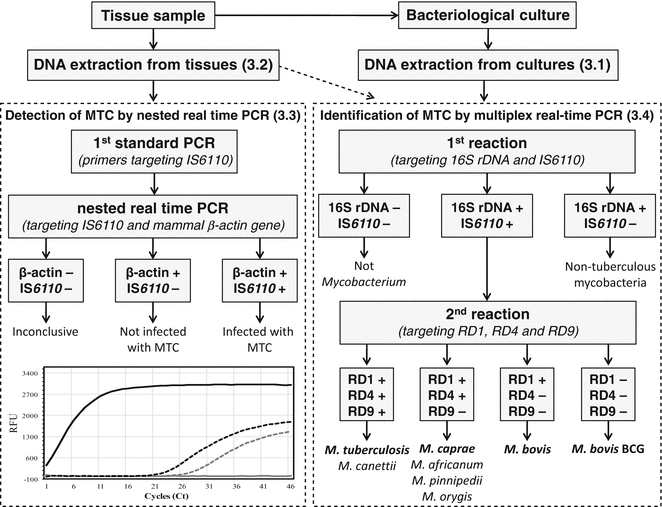

Fig. 1




Schematics of the detection and identification algorithm for MTC species most commonly associated with TB in livestock and other animals. DNA directly extracted from tissues can be used as template for the detection of MTC by nested real-time PCR targeting the IS6110 (left dashed box). The amplification of the mammal β-actin gene is used as control for the occurrence of inhibitors of the reactions. The inset in the bottom illustrates the amplification results usually obtained by the nested duplex real-time PCR assay of an MTC-infected (black lines) and non-infected (gray lines) tissues samples for the amplification targeting the IS6110 (solid lines) and the mammal β-actin gene (dashed lines) (RFU-relative fluorescence units). DNA extracted from cultures can be used as template for the identification of MTC by multiplex real-time PCR (right dashed box). The DNA extracted directly from tissues can be also used as template for these multiplex assays, but the sensitivity for the detection of MTC is lower than when using the nested real-time PCR assay. In the first amplification step, the isolate will be assigned as an MTC member (by detecting the presence of the IS6110 element) or, alternatively, as a non-MTC Mycobacterium species. The subsequent RDs-targeted triplex PCR allows the identification of the most veterinary-relevant MTC members to the species level as M. bovis (or M. bovis BCG), M. caprae, or M. tuberculosis, according to their distinct patterns of presence or absence of RD1, RD4, and RD9. Other MTC members such as “M. canettii,” M. africanum, M. pinnipedii, and M. orygis may present similar RDs profiles, but these species are rarely found in domestic (particularly livestock) and big game animals. Due to specific deletions spanning at least part of the region RD1, other less frequently found MTC species of M. microti and M. mungi present the alternative profile RD1 (−), RD4 (+), and RD9 (−)
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


