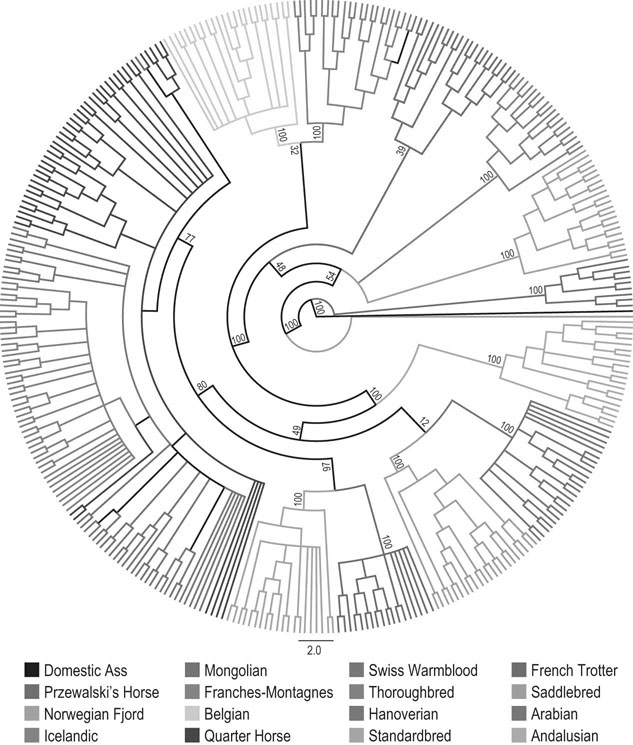
Equine breeding selection has recently been developed for some breeds and uses of horses by applying quantitative genetic methods for calculating the heritability of the complex traits, such as performance in racing or sport competitions. Genetic indices are now calculated routinely in different horse breeds in order to select the best stallions and mares according to a breeding plan with a well-defined genetic objective. Other exercise variables and disease characteristics have been studied by the same type of quantitative genetic methods. With development of biotechnologies, equine molecular genetics has come of age. The equine mitochondrial genome was sequenced in 19941 and then partial genome sequencing was undertaken.2–4 The recent sequencing of the entire equine genome by an international consortium5 was the major genetic result that will impact all the other equine genomics in the near future. With the rapid progress in equine genetics, new applications in early evaluation of the potential for performance and the detection of disease markers are available. Many new biomolecular tools will change the way selection, disease diagnosis and choice of treatment in athletic horses are made. In human exercise physiology, the research project HERITAGE family study provided continuous new knowledge and data about the genes and quantitative trait loci (QTL) related to exercise activity, health and performance. The last update of the gene list validated by experimental studies revealed 214 autosomal genes and quantitative trait loci and seven others on the X chromosome.6 Moreover, there are 18 mitochondrial genes that influence fitness and performance phenotypes. This important collection of genetic data related to exercise in humans demonstrates the great interest of studying equine genetics for performance evaluation and sports medicine applications. Thus, heritability (h2) is given by: Heritability values (h2) of 0.4 indicate that 40% of the trait could be transmitted to the offspring by the mare and stallion. Forty percent represents a high percentage of genetic influence. Note that heritability estimates are valid only for the breed for which they were calculated and they should be recalculated periodically if there is an active breeding program going in the population. The heritabilities of the performance traits are used to calculate genetic index using best linear unbiased predictor method (BLUP) in sport horses in different countries7. Performance heritability estimates in racing and equestrian sports are: 0.15 to 0.55 for flat gallop racing, 0.28 for endurance racing, 0.17 to 0.26 for trot or pace racing, 0.05 to 0.28 for show jumping and three-day-eventing and 0.11 for dressage.7–10 In addition, it was interesting to note that there is no or a weak negative genetic correlation between jumping ability and other sport performance. In show jumping, the longevity of the jumpers’ career could be considered as one important trait of the performance and its heritability is rather low 0.10.11 It has been studied in a large population of French saddle horses. Two main results were demonstrated: the younger the horse started competing, the longer it stayed in competition; the less success in competition, the greater was the chance for leaving competition, especially for horses without earnings. The risk to be removed from the competition is 1.45-fold higher for female than gelding, likely because of the use of females in breeding. Myosin Heavy Chain percentages were measured in small muscle samples of 20–30 mg collected by microbiopsy under local anesthesia in Anglo-Arabian horses. The analysis of slow MHC I and fast MHC IIA and IIX was performed using monoclonal antibodies with an ELISA method.12 The heritability of the percentage of fast MHC was 0.13 in Anglo-Arabian and 0.28 in Andalusian horses.13 The relative area h2 of the fast muscle fibres was 0.12 in Anglo-Arabian and 0.15–0.23 in Andalusian horses for fast and slow fibers, respectively. The capillary density heritability in Andalusian horses was 0.20. Their muscular trait heritabilities are rather low in comparison with Andalusian horses. The low heritabilies mean that the trait is genetically homogenous in the Anglo-arabian breed probably because the trait has been already selected for fast speed. The breeding selection on the muscular trait would be poorly efficient in this case. A group of more than 400 French trotters performed an exercise test at increasing speed. The heart rate and blood lactate were measured in order to determine the speed related changes of these variables. The speed eliciting 200 heart beats/min (V200) and a blood lactate concentration of 4 mmol/l (VLA4) were calculated. The V200 heritability was higher (0.46) than the VLA4 heritability (0.10) which was more influenced by the training effect.14 For harness trotters, the VLA4 is less heritable but more correlated to performance than V200.15 Breeding selection on V200 could be efficient to improve the sub-maximal heart rate capacity and thus aerobic capacity however the racing performance depends mainly on the ability to get a positive adaptation to training stimulation quantified by the VLA4 measurements. To measure kinetic and temporal variables of the walk, trot, gallop and jump the horses were equipped with the gait analysis system Equimetrix®.16,17 A three axial accelerometer was attached with an elastic girth onto the sternum. This ensured that the transducer was held close to the horse’s centre of gravity. The device recorded the dorso-ventral, medio-lateral and longitudinal acceleration of the horse during the exercise (Fig. 4.1). Gait tests in hand and free canter and jump tests provided gait variables which were used to calculate heritabilities in French saddle horses (Table 4.1 and Table 4.2). Heritabilities of the slow gaits such as walk and trot were lower that fast gaits such as canter and gallop. Heritabilities of canter, gallop and jumping variables, as with take-off impulse were rather high: 0.23 to 0.52.18 For equestrian sport horses, the use of gait variables such as stride frequency, propulsion power and jump variables could be efficiently used both for early selection and breeding selection. Table 4.1 Heritability (h2) of the gait variables measured in French saddle horses. The walk and trot were recorded in hand and the canter under free condition. Heritability estimates for canter were higher than for walk and trot h2: Heritability SE: standard error. NA: Non available data Table 4.2 Heritabilities (h2) of jump variables measured in French saddle horses and saddle pony. The jumps were recorded under free condition. Heritability for jump variables were higher than for walk or trot Using a two-dimensional image analysis, it has been possible to measure French saddle horses and to calculate the heritability estimates of the main morphological traits. Conformation is poorly heritable in Selle Français horses and more heritable in Poney de Selle.19 In the two breeds these conformation traits are poorly correlated to sport performance. However, in a review of the conformation heritabilties obtained by other methods (judge scoring and direct measurements) and in other breeds, the heritabilities are higher for height at withers (h2 = 0.25–0.89) or body weight (h2 = 0.13–0.90).20 The equine genome has been sequenced and annotated by an international consortium.5 The task has been conducted by the Broad Institute Sequencing Platform (Cambridge, MA, USA). All the database is available online at: http://www.ncbi.nlm.nih.gov/genome/guide/horse/. Briefly, the equine genome includes 20 322 predicted protein-coding genes according to the annotation process made by alignments and comparisons with the human, mouse and dog genome. Transcriptomic analysis showed that 87% of 18 039 genes can be detected using expression microarrays designed with equine probes. In order to have markers along all the equine genome, a single nucleotide polymorphism (SNP) map was designed including a total 106 SNP candidates with an average density of 1 SNP/2 kilo base. A first genotyping array including 54K SNPs was designed and validated by using a panel of 14 horse breeds, Equus Przewalski and other related Perissodactyl species (zebras, asses, tapirs, and rhinoceros)21. It was possible to discriminate between 14 breeds by genotyping a set of 1007 SNP (Fig. 4.2). These important results allows one to produce new genetic investigations in various domains and to design new genomic tools such as single nucleotide polymorphism chips for genotyping studies and expression microarrays to analyze gene expression. Gene expression and regulation determine most of the phenotype. The analysis of the cell transcriptome using DNA microarray provides qualitative and quantitative information about the up- and down-regulated pathways involved during exercise. The collected data are massive and complex and should be studied by sophisticated data mining methods in order to extract only the informative data. The first transcriptomic studies in exercising horses were performed in the blood leucocytes of performers and disqualified endurance horses22 and in muscles of Thoroughbred racehorses in training.23 The main results of these two studies are described in the following parts. Exercise stimulates immediate early molecular responses as well as delayed responses during recovery, resulting in a return to homeostasis and enabling long-term adaptation.23 Global gene expression occurring in equine skeletal muscle after exercise was explored using equine-specific cDNA microarrays to examine global mRNA expression in skeletal muscle from a cohort of eight Thoroughbred horses at three time points (before exercise, immediately afterexercise, and four hours after exercise) after a single bout of treadmill exercise. Skeletal muscle biopsies were taken from the gluteus medius before exercise, immediately after and four hours after exercise. While only two genes had increased expression after exercise (P < 0.05), by four hours after exercise 932 genes had increased (P < 0.05) and 562 genes had decreased expression (P < 0.05). Functional analysis of genes differentially expressed during the recovery phase (4 hours after exercise) revealed an over-representation of genes localized to the actin cytoskeleton and with functions in the MAPK signalling, focal adhesion, insulin signalling, mTOR signaling, p53 signalling and Type II diabetes mellitus pathways. After the exercise, there was an over-representation of genes involved in the stress response, metabolism and intracellular signalling. These findings suggest that protein synthesis, mechano-sensation and muscle remodeling contribute to skeletal muscle adaptation towards improved integrity and hypertrophy. In order to assess the short term response to exercise at the muscular level of three-year-old Thoroughbreds, the gene expression of creatine kinase (CKM), two sub-units of the respiratory chain complex IV (cytochrome c oxidase subunit IV isoform 1 (COX4I1), cytochrome c oxidase subunit IV isoform 2 (COX4I2) and pyruvate dehydrogenase kinase (PDK4) were measured by quantitative real time PCR after exercise.24 During high intensity sprint exercise, pyruvate dehydrogenase kinase (PDK4) was highly up-regulated in the gluteus medius muscle while during moderate intensity exercise on treadmill, one sub-unit of the respiratory complex IV was slightly down-regulated (COX4I2). PDK4 encodes a protein located in the matrix of the mitochondria and inhibits the pyruvate dehydrogenase complex by phosphorylating one of its subunits, thereby contributing to limit the glucose importation in mitochondria. Thus, PDK4 up-regulation after exercise increases beta-oxidation of fatty acids to acetyl-CoA as substrate for oxidative phosphorylation. This way, the pool of ATP can be efficiently restored. In a wide genome scan study of 112 Thoroughbred horses, some gene candidates related racing performance were identified by using 394 microsatellite markers.25 These genes were close to the genetic markers and were mainly involved in the energetic metabolism and muscle strength and integrity. The genes identified were in the energetic pathway (ADHFE1, alcohol dehydrogenase; PDK4 pyruvate dehydrogenase kinase 4), mitochondrial respiration (MTFR1, mitochondrial fission regulator 1; COX cytochrome units), hypoxia (HIF1A), fatty acid oxidation (ACSS1, acyl-CoA synthetase short-chain family member 1), phosphoinositide-mediated signalling, insulin receptor signalling (PI3KR1), and muscular proteins (ACTA1, ACTN2, TNC).
Genetic basis of equine performance
Introduction
Genomics of performance
Genetic component of the performance traits: how to measure the heritability of the exercise variables and performance traits using phenotypic measurements
Heritability calculation in horse performance
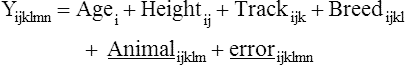
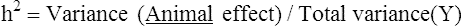
Muscle power and fiber composition in gluteus medius muscle
Evaluation of cardiac and aerobic capacity
Gait and jump tests
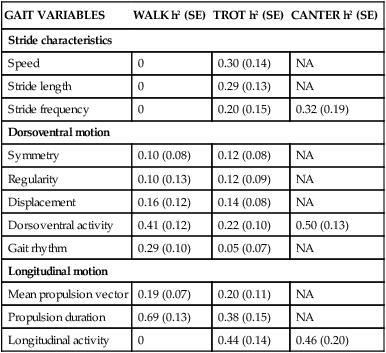
GAIT VARIABLES
WALK h2 (SE)
TROT h2 (SE)
CANTER h2 (SE)
Stride characteristics
Speed
0
0.30 (0.14)
NA
Stride length
0
0.29 (0.13)
NA
Stride frequency
0
0.20 (0.15)
0.32 (0.19)
Dorsoventral motion
Symmetry
0.10 (0.08)
0.12 (0.08)
NA
Regularity
0.10 (0.13)
0.12 (0.09)
NA
Displacement
0.16 (0.12)
0.14 (0.08)
NA
Dorsoventral activity
0.41 (0.12)
0.22 (0.10)
0.50 (0.13)
Gait rhythm
0.29 (0.10)
0.05 (0.07)
NA
Longitudinal motion
Mean propulsion vector
0.19 (0.07)
0.20 (0.11)
NA
Propulsion duration
0.69 (0.13)
0.38 (0.15)
NA
Longitudinal activity
0
0.44 (0.14)
0.46 (0.20)
TRAITS
HORSE
PONY
Approach strides
0.34
0.41
Take off force
0.23
0.43
Jump flight
0.30
0.42
Landing force
0.52
0.40
Conformation
Equine genetics and genomics come of age
Genomic applications in gallop and trot racing
Short time genomic response to exercise in Thoroughbreds
Genotyping in Thoroughbreds for gallop racing ability
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Genetic basis of equine performance
