Chapter 5
Clinical Virology
A number of viruses have been shown to be important pathogens in reptiles. In other cases, the association between viral infection and disease is not clear, and the discovery of some viruses in reptiles has been incidental. Reptile virology is still a relatively young field and much remains to be discovered and understood. The clinical significance of viral infections appears to depend on many different factors including the virus family, the virus strain, the host species, factors affecting the host immune system, and additional infectious agents such as other viruses, bacteria, and parasites. Interpretation of diagnostic testing requires an understanding of the difference between infection and disease. Infection with a virus simply means that the virus has invaded and replicated in the body of the host. This may or may not lead to the development of disease. We are still in the process of understanding which viruses can be found in reptiles. It is even more difficult to interpret their clinical significance and to develop methods to detect them in infected reptiles, as well as deal with infected animals. This review will concentrate on the laboratory diagnosis of common viruses found in reptiles and possible interpretation of laboratory results. It is not meant as an overview of all viruses described in reptiles (an overview is provided in Appendix 2. See Jacobson1 and Marschang2 for comprehensive reviews of viruses detected in reptiles).
Viruses of Chelonians
A number of viruses have been described in chelonians. Among this group of animals, the viruses most commonly detected have been herpesviruses (HVs), which have been associated with various disease syndromes but have also been found in apparently healthy animals in some cases. HVs have been described most commonly in chelonians of the families Testudinidae (tortoises) and Cheloniidae (sea turtles). Other viruses that have been regularly detected in this group of animals are ranaviruses and picornaviruses. Individual reports have been published on the detection of adenoviruses (AdVs), PMVs, reoviruses, and papillomaviruses in chelonians. A list of viruses regularly detected in chelonians and methods for diagnostic testing can be found in Table 5-1.
Herpesviruses
HVs are enveloped, double-stranded (ds) deoxyribonucleic (DNA) viruses. These viruses are relatively susceptible to disinfectants and are therefore easy to inactivate in the environment with the use of standard virucidal disinfectants. However, HV infections lead to lifelong latency. Latent virus remains inactive in infected cells—in the case of alphaherpesviruses, in neuronal cells. The virus can then begin to replicate again under some circumstances; thus an HV-infected animal that survives initial infection must be considered a lifelong carrier. HVs have been detected in many different species of turtles and tortoises as well as other groups of reptiles. In chelonians, HVs have been described in water turtles (Emydidae), tortoises, and sea turtles. In water turtles, the presence of HVs has been based on histologic detection of intranuclear inclusions and electron microscopy of infected tissues. Infections were associated with hepatic disease.3–5
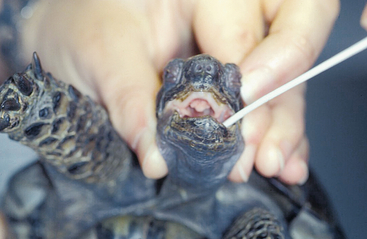
In sea turtles, HV infections have been associated with skin lesions (gray patch disease), fibropapillomatosis, lung, eye, and trachea disease (LETD), loggerhead genital–respiratory HV (LGRV)-associated disease, and loggerhead orocutaneous HV (LOCV)-associated disease. Gray patch disease was the first HV-associated disease to be described in sea turtles and was found in aquaculture-reared young Green Sea Turtles (Chelonia mydas).6 Another HV-associated disease described in Green Sea Turtles was characterized by gasping, harsh respiratory sounds, buoyancy abnormalities, inability to dive properly, and the presence of caseated material on the eyes, around the glottis, and within the trachea. This disease was named LETD.7 Two HV-associated disease syndromes have been described in wild-caught Loggerhead Sea Turtles (Caretta caretta). Turtles infected with LGRV had ulcers in the trachea, around the cloaca, and on the base of the phallus. Turtles infected with LOCV had pneumonia and ulcers and cutaneous plaques covered with exudate in the oral cavity.8 The most commonly described HV-associated disease in sea turtles is fibropapillomatosis (Figure 5-1). Fibropapillomatosis has been detected in Green, Loggerhead, Hawksbill (Eretmochelys imbricata), and Olive Ridley (Lepidochelys olivacea) Sea Turtles around the world. Infected turtles develop fibropapillomas, and individual or multiple tumors can occur externally all over the body. Internal tumors are also possible. Although the fibropapillomatosis HV has never been isolated in cell culture, transmission of the disease is possible with the use of cell-free tumor extracts.9
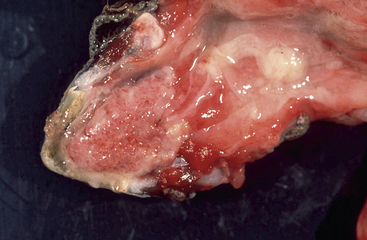
In tortoises, HV infections have mostly been associated with diphtheroid-necrotizing stomatitis (Figure 5-2). This can be quite severe and often leads to the death of the affected animal. Rhinitis, conjunctivitis, cervical edema, anorexia, and lethargy are also frequently observed. Central nervous disorders such as paralysis and incoordination, as well as hepatitis, have also been described. Eosinophilic or amphophilic intranuclear inclusions are often detected in infected tissues, most commonly in epithelial cells of the tongue, oral mucosa, and upper respiratory tract as well as the gastrointestinal tract (Figure 5-3). Other tissues in which inclusions have been described include the urinary tract, the brain, the liver, and the spleen. In some cases, HV infections have also been detected in clinically healthy tortoises. Development of disease and prognosis appear to depend both on the host species and on the virus involved.10 There are currently four different HVs that have been shown to infect tortoises. They have been named testudinid HV 1 to 4 (TeHV1 to 4).2 TeHV1 was first detected in Russian Tortoises (Testudo horsfieldii) and Pancake Tortoises (Malacochersus tornieri) in Japan.11 Similar viruses have also been found in tortoises in Europe. Although they have been detected in several different species, all cases so far have had direct contact with Russian Tortoises. TeHV1 is associated with stomatitis in infected animals but does not appear to cause high morbidity or mortality. TeHV2 was described in a California Desert Tortoise (Gopherus agassizii) in the United States. The animal exhibited anorexia, lethargy, and yellow-white caseous plaques on the tongue and palate.12 TeHV3 has been most frequently described in Mediterranean tortoises (Hermann’s [T. hermanni], Spur-thighed [T. graeca] and Marginated [T. marginata] Tortoises) and Russian Tortoises in Europe. It has also been detected in tortoises in the United States and in northern Africa. This virus is associated with severe disease and high morbidity and mortality, particularly in Hermann’s and Russian Tortoises. Spur-thighed Tortoises develop disease less frequently and appear to be able to survive and carry the infection.13 TeHV4 was detected in a clinically healthy Bowsprit Tortoise (Chersina angulata) in a zoo in the United States.14
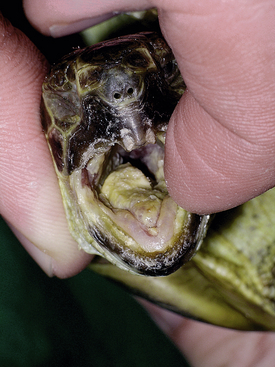
FIGURE 5-2 Russian Tortoise (Testudo horsfieldii) with herpesvirus infection. Severe stomatitis with diphtheroid plaques visible throughout the oral cavity. (Photo courtesy of Dr. Volker Schmidt, Universität Leipzig, Leipzig, Germany.)
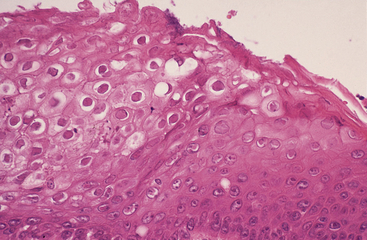
FIGURE 5-3 Herpesvirus infection in a tortoise (Testudo hermanni). Photomicrograph of the epithelium of the tongue. Ballooning degeneration is evident in numerous epithelial cells. Large intranuclear inclusions are also visible in several cells. H&E stain, × 400. (Photo courtesy Dr. Horst Posthaus, Universität Bern, Berne, Switzerland.)
Diagnosis of Herpes Virus Infections in Chelonians
Virus Detection
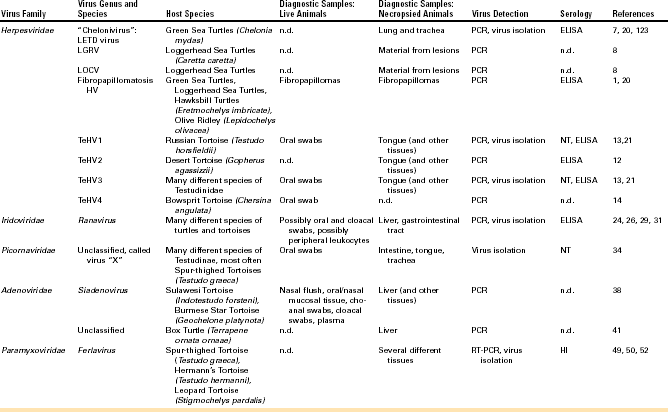
Detection of viral DNA by PCR is the most commonly used method for detecting HVs in infected chelonians. A PCR using degenerate primers in a nested format targeting a highly conserved portion of the DNA polymerase gene has been used to detect HVs in many different chelonian species.8,13,15 This method has been shown to be the most sensitive available for HV detection in tortoises. Other PCRs have been described targeting specific TeHVs.16–18 All of these methods are, however, less sensitive than the nested PCR but more specific. This means that, although the nested PCR can detect all of the chelonian HVs tested so far, other methods are limited to one or two different virus species (e.g., only TeHV1 or only TeHV3), and other virus species will not be detected if these methods are used. Virus detection by PCR should be verified by additional methods (e.g., sequencing of the PCR product, especially when a PCR with degenerate primers is used). Virus isolation in cell culture has also been used to detect HVs in infected chelonians. LETD virus (LETV), TeHV1, and TeHV3 have all been isolated in cell culture (Figure 5-4, B). Isolation in cell culture is generally less sensitive than the nested PCR already described. It is time consuming and is offered by very few laboratories. It can be helpful for the detection of viruses other than HVs (e.g., picornaviruses; see further on), which may be important differential diagnoses for herpesviral disease or may be involved in disease processes. Samples for HV detection in chelonians should generally include tissues with lesions. For fibropapillomatosis, viral DNA can be detected in fibropapillomas removed from live or dead animals. In tortoises, HV DNA has been detected in oral swabs from live animals. Swabs should be taken from the base of the tongue and should include cellular material (Figure 5-5). In dead tortoises, the tongue is generally considered the best tissue for virus detection. Esophagus, stomach, intestine, trachea, liver, and brain can also be helpful in virus detection.
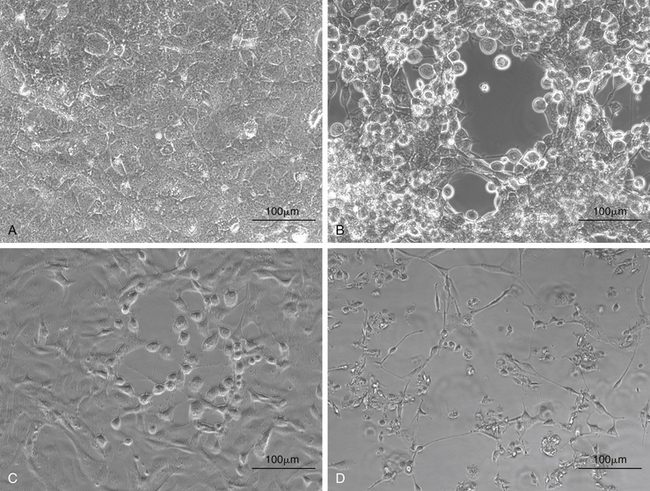
FIGURE 5-4 Isolation of chelonian viruses in cell culture. A, Uninfected terrapin heart cells (TH1), which are often used for the isolation of chelonid viruses. B, TH-1 infected with a testudinid herpesvirus 3 (TeHV3). This virus causes a cytopathic effect (CPE) with cell lysis and rounding of infected cells. C, TH1 infected with a ranavirus isolated from a Hermann’s tortoise (Testudo hermanni). Ranaviruses from reptiles, amphibians and fish grow in a wide range of cell lines and cause CPEs with cell lysis and rounding of infected cells. D, TH1 infected with the picornavirus (virus “X”). This virus causes a CPE with cell lysis.
Serology
Detection of antibodies against HVs is particularly important because of the biologic properties of these viruses. Because HVs cause latent infections, any animal found to be serologically positive for HVs must be considered a lifelong carrier, even if the animal appears healthy. Serologic tests have been described for the detection of antibodies against fibropapillomatosis-associated HV and LETV in sea turtles and against TeHV1 and TeHV3 in tortoises.19–21 An ELISA for the detection of antibodies against glycoprotein H of a chelonid fibropapillomatosis-associated HV has been described.19 High seroprevalences were found in wild Green Sea Turtles in Florida and in Loggerhead Turtles using this method. Seropositivity did not correlate with clinical disease. However, testing for antibodies against this virus is not widely available.
An ELISA has also been described for the detection of antibodies against LETV. This ELISA was specific for LETV and did not detect antibodies against fibropapillomatosis-associated HV.20 Virus neutralization tests and ELISAs have been described for the detection of antibodies against TeHV1 and against TeHV3. Virus neutralization tests are most commonly used in Europe, whereas ELISAs are used in the United States. Virus must be able to grow in cell culture for the development of a virus neutralization test. Virus neutralization testing with both TeHV1 and TeHV3 has shown that these viruses do not cross-react serologically; thus testing for antibodies against both is recommended in Europe. Detection of antibodies against these two viruses has been shown to depend on the tortoise species involved. Hermann’s Tortoises, which are particularly susceptible to HV infection and disease, do not often develop neutralizing antibodies after infection. In contrast, antibodies are frequently detected in Spur-thighed Tortoises that have been infected. An ELISA has also been developed for the detection of antibodies against TeHV3 in tortoises.21 This ELISA has also been adapted to detect antibodies against TeHV2 in California Desert Tortoises, on the basis of putative serologic cross-reactivity between TeHV2 and TeHV3.12,22
Ranaviruses
Ranaviruses belong to the family Iridoviridae, a group of large, dsDNA viruses infecting only ectothermic hosts including various invertebrates, fish, amphibians, and reptiles. Ranaviruses are enveloped but do not need their envelope to be infectious. In reptiles, ranaviruses have most commonly been detected in chelonians, although they have also been described in snakes and lizards. In chelonians, these viruses have been found in Russian Tortoises, Eastern Box Turtles (Terrapene carolina carolina), Chinese Soft-shelled Turtles (Pelodiscus sinensis), Hermann’s tortoises, Red-eared Sliders (Trachemys scripta elegans), Burmese Star Tortoises (Geochelone platynota), Gopher Tortoises (Gopherus polyphemus), Florida Box Turtles (Terrapene carolina bauri), Egyptian Tortoises (Testudo kleinmanni), a Leopard Tortoise (Stigmochelys pardalis), Marginated Tortoises, and Spur-thighed Tortoises.23–30 Ranavirus infection in chelonians has been associated with lethargy, anorexia, nasal discharge, conjunctivitis, severe subcutaneous cervical edema, ulcerative stomatitis, and “red-neck disease” (Figure 5-6). Histologically, infected animals have been found to have hepatitis, enteritis, and pneumonia. Infected cells, particularly epithelial cells of the gastrointestinal tract and hepatocytes, may have basophilic intracytoplasmic inclusions. In a transmission study with Box Turtles (Terrapene ornata ornata) and Red-eared Sliders, intramuscular injection of a ranavirus isolated from a Burmese Star Tortoise led to disease, including lethargy, anorexia, ocular discharge, conjunctivitis, and oral plaques, and death of the animals in some cases.24 Ranaviruses do not appear to be species specific and can be transmitted between animal species and possibly even between different classes of animals.
Diagnosis of Ranavirus Infections in Chelonians
Ranaviruses grow well in cell culture and can be grown on a wide range of cell lines from reptiles, fish, mammals, and birds if the cells are kept at appropriate temperatures (below 32°C or 89.6°F) (see Figure 5-4, C). Virus isolation on cell culture is a sensitive method for the detection of ranaviruses. PCRs for the detection of viral DNA have also been used to diagnose ranavirus infections in chelonians. The gene most frequently targeted is the major capsid protein (MCP) gene.23,26 This gene is highly conserved in ranaviruses and is therefore a good target for virus detection. PCR-positive reactions are generally specific, but confirmation of product identity by sequencing is recommended. Other PCRs, including real-time PCRs, have been described for the detection of ranaviruses in amphibians and fish but have not yet been used for ranavirus detection in chelonians. Samples for the detection of ranaviruses in dead chelonians should include liver and gastrointestinal tract. Spleen and kidney may also be virus positive.24 In a transmission study viral DNA was detected in oral and cloacal swabs from intramuscularly infected Red-eared Sliders as early as 5 days postinoculation (p.i.) and until 26 days p.i. or until the animals died or were euthanized.24 Both oral swabs and blood have been used to detect ranavirus in free-ranging Eastern Box Turtles in the United States, whereas oral and cloacal swabs have been used for the detection of viral DNA in naturally infected tortoises in Germany.29,31
Serology
Little work has been done studying the immune response of reptiles to ranaviral infection. An ELISA for the detection of anti ranavirus antibodies in Burmese Star Tortoises, Gopher Tortoises, and Eastern Box Turtles has been developed.32 The assay is able to detect antibodies in all three species. Testing of 1000 Gopher Tortoises from the eastern United States showed a low prevalence of anti ranavirus antibodies of 1.5% overall, whereas prevalence in a group of 55 Eastern Box Turtles that had survived an outbreak of ranavirus infection 1 year previously was 1.8% (one positive). The authors postulated that the low prevalence rates detected may underestimate the true prevalence of infection, as ranavirus infections in chelonians are often associated with high mortality rates and the antibody response is unknown.32 This ELISA has also been adapted for the detection of anti ranavirus antibodies in Testudo spp. in Europe.29
Picornaviruses
Picornalike viruses are frequently found in tortoises in Europe. The family Picornaviridae contains small single-stranded (ss), nonenveloped ribonucleic acid (RNA) viruses that can be relatively resistant to disinfection and can persist for extended periods of time in the environment. In tortoises, picornalike viruses have been detected by isolation in cell culture with the use of chelonian cell lines. Because of difficulties in characterizing these viruses, they have been called virus “X”.33 These viruses have most frequently been isolated from Spur-thighed Tortoises. They are also found in Marginated Tortoises, Hermann’s Tortoises, Leopard Tortoises, and Egyptian Tortoises. Clinical signs associated with infection include softening of the carapace in young animals, diphtheroid-necrotizing stomatitis, rhinitis, conjunctivitis, and ascites (Figure 5-7). Viruses have also been isolated from clinically healthy animals34 (Heuser: Pers. com.). There are no typical histologic lesions associated with infection. Many tortoises infected with virus “X” are also infected with other infectious agents, including HVs and Mycoplasma spp. (Marschang RE, unpublished observations). A large part of the genome of one virus “X” isolate has recently been almost fully sequenced, showing that this is a novel picornavirus.35,36 The sequence information now available will allow the development of new diagnostic tools, including reverse transcriptase PCR (RT-PCR) as well as the comparison of virus “X” isolates from different tortoise species, different geographic locations, and different years.
Diagnosis of Picornaviruses in Tortoises
The detection of virus “X”-like viruses in tortoises currently relies on isolation in cell culture. Virus can be isolated on TH-1 (American Type Culture Collection [ATCC] No. CCL-50) at 28°C (82.4°F). The virus causes a lytic cytopathic effect in infected cells (see Figure 5-4, D). Virus identification after isolation is difficult and relies on the typical cytopathic effect (CPE) and resistance to chloroform. Visualization of virions in infected cell cultures by electron microscopy is difficult. Although no RT-PCR is currently available for the detection of these viruses in clinical samples, primers are available that can be used to help identify the viruses after isolation in cell culture. PCR products can then be sequenced and compared with available sequence data. The best samples for virus detection in live animals are oral swabs. Virus can also be shed via the cloaca and the conjunctiva, so swabs from these areas may also yield positive results. In dead animals, samples from the entire gastrointestinal tract (tongue to cloaca) can all be used for virus detection. Virus is frequently also found in other tissues (including liver, kidney, heart, brain, and lung). Serologic detection of antibodies against virus “X” can be carried out using virus neutralization methods. Antibodies against this virus are frequently found in tortoises in Europe, and low titers have also been detected in wild-caught tortoises in Turkey.37 It is unknown how closely related all virus “X” isolates are to one another, so cross-reactivity among these viruses and between these viruses and other picornaviruses is not yet understood.
Other Viruses Described in Chelonians
Adenoviruses
AdVs are nonenveloped, dsDNA viruses that have a relatively high resistance to inactivation and can be difficult to disinfect. In reptiles, they are most commonly detected in squamates, particularly agamids, but AdVs have recently also been described in several species of chelonians including Sulawesi Tortoises (Indotestudo forsteni),38 Impressed Tortoises (Manouria impressa), a Burmese Star Tortoise,39 a Leopard Tortoise40, and a Box Turtle.41 Infection in the Sulawesi Tortoises was associated with severe systemic disease and a mortality rate of 82%. Pathologic findings in infected tortoises were multifocal hepatic necrosis, amphophilic to basophilic intranuclear inclusions and diffuse hepatic lipidosis, myeloid necrosis in bone marrow, and severe necrotizing enterocolitis.38 The Impressed Tortoises and the Burmese Star Tortoise were all exposed to infected Sulawesi Tortoises.39 The Leopard Tortoise was also infected with a herpesvirus and had biliverdinuria, wasting, and episodes of hemorrhages.40 The infected Box Turtle showed liver changes with cellular degeneration, pronounced vacuolization of the cytoplasm and pyknosis of nuclei, and inclusion bodies in some hepatocytes.41 Diagnosis of AdVs is generally carried out with the use of PCR targeting a portion of the DNA polymerase gene.42 This PCR uses degenerate primers and targets a highly conserved region of the genome and can therefore be used to detect a wide range of AdVs from different hosts. PCR products should therefore be sequenced to confirm the identity of the virus detected. In live animals, nasal flushes, oral/nasal mucosal tissue, choanal swabs, cloacal swabs, and plasma have all been successfully used to detect virus.38 Liver tissue has been successfully used for virus detection in dead animals,38,41 as have a number of other internal tissues.38 The AdVs detected in chelonians so far do not appear to be closely related to the AdVs of squamates, and several appear to differ significantly from one another. No serologic test is currently available for the detection of antibodies against AdVs in chelonians.
Papillomaviruses
Papillomaviruses are nonenveloped dsDNA viruses with a circular genome. The papillomaviruses are highly host–species specific and tissue restricted. They generally cause benign tumors (warts, papillomas) in their natural hosts. Occasionally, they can also cause these lesions in related species. Papillomaviruses are highly resistant to inactivation and can persist in the environment for long periods of time. In chelonians, papillomaviruses have been described in Bolivian Side-neck Turtles (Platemys platycephala)43, a Russian Tortoise,44 a Loggerhead Turtle, and a Green Sea Turtle.45 Skin lesions were reported in all but the Russian Tortoise, which had a history of stomatitis. In that case, papillomavirus-like particles were detected by electron microscopy in a lung wash but not in oral scrapings.44 Diagnosis of papillomavirus infections has been mostly by detection of viral particles in infected tissues via electron microscopy. The complete sequences of the papillomaviruses of the Loggerhead and Green Sea Turtles have been determined, so future development of a diagnostic PCR is possible.46 Tissues for analysis should include affected skin. Papillomaviruses do not generally grow well in cell culture, and no reptilian papillomavirus has been isolated. There is no serologic test available for the detection of antibodies against these viruses in chelonians.
Reoviruses
Reoviruses are nonenveloped RNA viruses with a dsRNA genome. There is only a single reported case of reovirus infection in a Spur-thighed Tortoise that was cachectic and had necrosis of the epithelium of the tongue. The virus was isolated from the tongue of that animal, as well as from several internal tissues.34 A virus neutralization test has been used to detect antibodies against this virus in tortoises.37
Paramyxoviruses
PMVs are enveloped, ssRNA viruses. In reptiles, these viruses are most commonly detected in snakes, but they have also been described in lizards and tortoises. Most of the PMVs detected in reptiles so far have been classified in the new genus Ferlavirus.47 In chelonians, PMV infections have been associated with dermatitis in a group of Spur-thighed Tortoises imported into Switzerland from Turkey.48 In a Hermann’s Tortoise and in a Leopard Tortoise, ferlavirus infection was associated with pneumonia. The Leopard Tortoise had been imported into Germany from Kenya several years previously and died with severe respiratory disease (Figure 5-8). A ferlavirus was detected in several internal tissues from that animal but not from the lung.49 A Hermann’s Tortoise is the only case in which a Ferlavirus from a tortoise has been isolated in cell culture so far.50 Diagnosis of ferlavirus infection in tortoises can be carried out with the use of an RT-PCR targeting the L gene of ferlaviruses similar to the process for snakes.51 This PCR often results in false-positive findings or products that are not the expected size and confirmation of positive results; therefore sequencing of PCR products is strongly recommended. Because ferlaviruses cause hemagglutination of chicken red blood cells, antibodies against these viruses in tortoises can be detected by HI assays, just as for snakes. Several different ferlaviruses have been detected in tortoises,49,50 so the choice of antigen may affect test results. However, a study has indicated that there is significant serologic cross-reactivity between known ferlaviruses.52

FIGURE 5-8 Leopard tortoise (Stigmochelys pardalis) with severe pneumonia. A ferlavirus was detected in this animal. (From Papp T, Seybold J, Marschang RE. Paramyxovirus infection in a leopard tortoise [Geochelone pardalis babcocki] with respiratory disease. J Herpetol Med Surg 2010;20:64-68. With permission.)
Viruses of Squamates
A variety of viruses have been described in lizards and snakes. Lizards are not monophyletic, so relationships between some lizards (e.g., iguanids) and snakes are closer than between other lizard families. This is reflected in the relationships between the viruses infecting some of these animals. PMVs are well-known pathogens in viperid snakes but can also be found in other families of snakes as well as in lizards. Both AdVs and reoviruses are regularly detected in lizards and snakes. A list of viruses regularly detected in squamates and methods for diagnostic testing can be found in Table 5-2.
Paramyxoviruses
PMVs are enveloped viruses with a negative sense ssRNA genome. They are relatively unstable in the environment. Almost all PMVs detected in reptiles are classified in the genus Ferlavirus,47 which contains PMVs detected in snakes, lizards, and chelonians. The ferlaviruses are named after the first PMV isolated from reptiles, Fer-de-Lance vipers in a Swiss serpentarium. During that outbreak in 1972, 87% of the snakes in one room died with dyspnea, opisthotonus, apathy followed by abnormal activity, mydriasis, and terminal convulsions.53 Since then, ferlavirus outbreaks have been documented in numerous snake collections in North and South America and Europe. Common clinical signs described in infected snakes include abnormal posturing, regurgitation, anorexia, head tremors, abnormal respiratory sounds, and exudate in the oral cavity (Figure 5-9). In many cases, no clinical signs may be noticed, and infected animals may be found dead in their enclosures.1,53,54 Severe disease has mostly been described in viperid snakes, but ferlaviruses have been found in snakes from the families Boidae, Elapidae, Colubridae, and Crotalidae.1 Koch’s postulates have been fulfilled for pulmonary lesions associated with ferlavirus infection in Aruba Island Rattlesnakes (Crotalus durissus unicolor). A ferlavirus isolated from several tissues of an Aruba Island Rattlesnake that died of the infection was inoculated intratracheally into four Aruba Island Rattlesnakes. Several snakes developed pulmonary symptoms including blood in the lungs, trachea, and oral cavity. The two animals that were not euthanized earlier died 19 and 22 days p.i.55 Gross abnormalities are most consistently found in the lungs of infected snakes. Changes include congestion and hemorrhage. Histologic findings often show proliferative interstitial pulmonary disease with proliferation and vacuolation of epithelial cells lining the faveoli. In rare cases, intracytoplasmic inclusions can be seen in lining epithelial cells.54,55 Although ferlaviruses are most commonly described in snakes, these viruses have also been detected in several species of lizard, including a Spotted False Monitor (Callopistes maculatus),56 an Emerald Tree Monitor (Varanus prasinus),57 a Flathead Knob-scaled Lizard (Xenosaurus platyceps),58 and a group of Caiman Lizards (Dracaena guianensis).59 Ferlavirus infections in lizards have been associated primarily with pneumonia, although virus infections in clinically healthy lizards have been documented. The viruses do not appear to be host specific, and transmission between different species of snakes and lizards as well as chelonians may be possible. There is no specific treatment available for ferlavirus infection in reptiles. The results of a single study attempting to vaccinate rattlesnakes against ferlavirus infection with the use of an inactivated cell culture isolate were equivocal, in that antibody responses were variable and transient.60

FIGURE 5-9 Bush Vipers (Atheris squamigera) infected with ferlaviruses. A, Dyspnea and bloody secretions in the oral cavity. B, Star gazing. (Courtesy of Jutta Wiechert.)
A PMV that differs distinctly from the ferlaviruses has recently been described in snakes in Australia. The virus has been preliminarily named Sunshine virus for the geographic location of the first isolation on the Sunshine coast of Australia. This virus was associated with neurorespiratory disease in Australian pythons.61
Diagnosis of PMV Infections in Squamates
Virus Detection
Ferlaviruses were first diagnosed in snakes with the use of isolation in cell culture followed by virus characterization.62 Virus isolation has since been used in many cases to detect these viruses in clinical samples from snakes and lizards. Ferlaviruses can be isolated on Russell’s Viper heart cells (VH2, ATCC, CCL-140) at 28°C (82.4°F) (Figure 5-10, A). They grow relatively slowly (7 to 14 days) and cause the formation of syncytia (Figure 5-10, B). A number of RT-PCRs have also been described for the detection of ferlaviruses in reptiles. The most sensitive test available to date is an RT-PCR targeting the large polymerase (L) gene.50,51 This gene is relatively well conserved among PMVs, and the primers described for this RT-PCR have been able to detect all of the ferlavirus types detected to date. This RT-PCR can be used to detect ferlaviruses in tissues from dead animals or in samples from live animals. In live animals, oral and cloacal swabs, as well as transtracheal washes, can be used as diagnostic samples. Because oral and cloacal shedding can be inconsistent, it is recommended that a combined sample of both be submitted, possibly combined with fluid from a transtracheal wash for testing.63 In dead animals, the highest viral load is found in the lung, and this tissue is recommended for testing.64 The RT-PCR is not highly specific for ferlaviruses, and false-positive reactions have been shown to occur. For this reason, PCR products of the expected size (566 base pairs) should be sequenced.
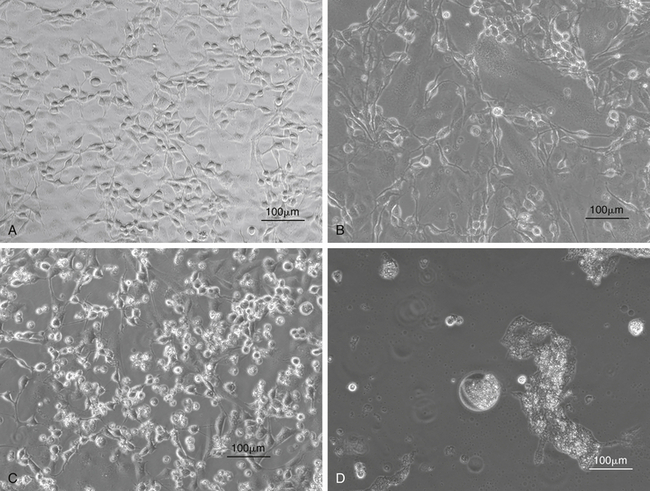
FIGURE 5-10 Isolation of snake viruses in cell culture. A, Uninfected viper heart cells (VH2), which are often used for the isolation of viruses from snakes. B, VH2 infected with a ferlavirus. This virus causes a cytopathic effect (CPE) with syncytia formation in infected cells. C, VH2 infected with an adenovirus isolated from a Boa Constrictor (Boa constrictor) (snake adenovirus 1). These viruses cause a CPE with cell lysis and rounding of cells. D, VH2 infected with a reovirus isolated from a Boa Constrictor. These viruses cause a CPE with the formation of giant syncytia.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree