Chapter 20 Approaching Health Problems at the Wildlife–Domestic Animal Interface
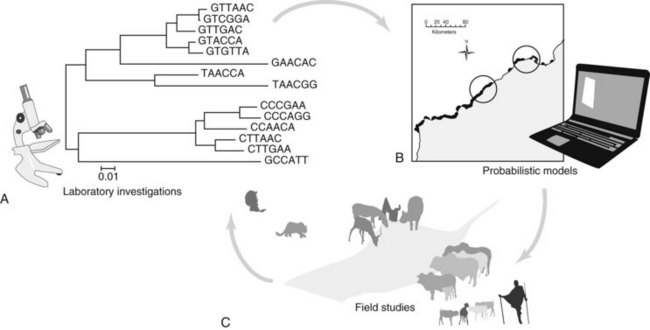
One solution to disease and conservation at the wild-domestic animal interface is the implementation of a proactive approach—addressing potential pathogen transmission and the resulting disease transmission before a volatile problem occurs. A wildlife disease surveillance and response strategy that is measurable, adaptive, and responsive is critical for detecting and monitoring cross-species disease transmission, especially zoonotic pathogens that account for most emerging infectious diseases in people, such as influenza and West Nile virus.15 In fact, more than 75% of these emerging zoonoses are the result of wildlife origin pathogens.6 Another useful tool for improving the efficiencies of surveillance efforts at wild-domestic animal disease hot spots is the use of important sentinel species. This approach facilitates the improvement of infectious disease models, resulting in higher quality field studies and more specifically targeted molecular diagnostics, and eventually in more effective conservation management strategies (Fig. 20-1).
Special Considerations at the Wild-Domestic Animal Interface
Unlike highly pathogenic, single-host pathogens that are often not sustainable in very small wild animal populations, multihost pathogens may be maintained by a reservoir host if there is recruitment of susceptible individuals in the population.8 Domestic animal populations, such as livestock herds and feral dog packs and cat colonies, may be able to maintain domestic animal pathogens that are infectious to wildlife and provide a continuous supply of immunologically naive young individuals. Domestic animal populations are capable of being infected with pathogens of wildlife origin if herd health is not maintained through good disease prevention practices. Therefore, domestic animal populations may be able to serve as a reservoir for wildlife pathogens. Similarly, wildlife populations may carry pathogens that do not cause clinical disease in the wild animal host but may cause significant problems to health and productivity in domestic animals at ecological interfaces likely to facilitate transmission. Thus, domestic and wild animal populations may be important sources of pathogens capable of crossing into nearby susceptible animal populations, resulting in spillover and spillback of pathogens of concern.
Approaching Novel Wildlife Disease Problems
For wildlife disease studies to result in justifiable and implementable interventions, we suggest a strategic combination of field studies to identify pathogens and their sources, molecular diagnostics for appropriate characterization, and dynamic modeling of the system and potential interventions using a modern risk-based approach. As depicted in Figure 20-1, the modeling helps target and improve field studies, and the data from field studies and molecular diagnostics iteratively improve the risk-based models and recommended interventions.
In many cases, a disease investigation will be the first scientific endeavor to characterize and address an emerging health problem affecting conservation. In these cases, a consistent and measurable process is useful for efficiently designing the approach without falling into a trap of following hunches or leads that may not be at the root of the problem. An example of an approach for comprehensive health studies at the wild-domestic animal interface that may be applied consistently is illustrated in Box 20-1. Slight variations of this approach have been applied successfully in multiple situations in developed and developing country contexts. The following sections provide specific examples for both—identification and mitigation of a disease threat in the threatened sea otter population in California, and the effect of water restriction on zoonotic disease transmission at the domestic-wild animal interface in the Ruaha ecosystem, Tanzania.
BOX 20-1 Ten Steps for Health Studies of Disease Emergence in Wild Animal Populations at Complex Interfaces* * This is an example of a consistent approach that may be useful in efficiently designing comprehensive health studies at the wild animal–domestic animal interface.
Multidisciplinary Sea Otter Research Strategy in California
Sea otters (Enhydra lutris) have been designated as a federally listed threatened species in California and Alaska because these subpopulations have shown limited or sporadic recovery since overhunting decimated their numbers along the Pacific Rim during the height of the fur trade in the 18th and 19th centuries. Infectious disease and limited prey resources are two key interacting factors impeding recovery of southern sea otters (Enhydra lutris neries) in California.5 Septic peritonitis caused by migration of acanthocephalan parasites and protozoal encephalitis caused by Sarcocystis neurona and Toxoplasma gondii have been reported as common infectious diseases detected in sea otter carcasses found beach-cast along the California coast.7,10,16
Toxoplasma gondii is a ubiquitous, apicomplexan, protozoal parasite with a complex multihost life cycle and an infective oocyst life stage that may persist in the environment. This pathogen may infect all warm-blooded vertebrates, including humans, but domestic and wild felids are the only known definitive hosts for T. gondii.3 The unusually high frequency of T. gondii exposure and infection in marine mammals has been a fascinating puzzle, considering the terrestrial nature of all recognized definitive hosts. A wide range of marine mammals has been reported with infections, and these species vary markedly in habitat and prey preference.9,12 Sea otters and most marine mammals do not consume known intermediate hosts for T. gondii. Whereas the processes promoting these infections in marine mammals are undoubtedly complex, T. gondii is a multihost pathogen widely recognized for its ability to spill over into susceptible wild animal populations, and it is likely that this pathogen has emerged in sea otters from cats and wild felids.2
As is most often necessary when initiating health investigations in wild animal populations, basic health screening was initiated for opportunistically sampled live, free-ranging, California and Alaskan sea otters to establish normal health parameter reference ranges and evaluate exposure to a host of possible pathogens that might cause disease in this species. Initial health assessments and baseline disease exposure information provides an historical context of past exposure patterns that may be useful for understanding future disease events. These efforts also identify potential disease issues and provide a focus for disease investigations. This initial effort in sea otters resulted in the first report of a high prevalence (35%) of T. gondii exposure among 77 California otters sampled between 1995 and 2000.4 Seroprevalence, calculated as the number of individuals with serologic evidence of past exposure to a pathogen divided by the number of individuals sampled, may provide insight into the frequency of pathogen exposure during a specified period of time, provided that adequate numbers of individuals are sampled that are representative of the overall population. It is often necessary to calculate age-specific seroprevalence for each age class sampled and evaluate differences by other population characteristics of interest to understand patterns in pathogen exposure better. Furthermore, patterns in pathogen exposure may change quickly, and it is critical to update measures of seroprevalence for pathogens of interest. Seroprevalence studies have identified geographic areas with high levels of T. gondii seroprevalence and determined that adult otters were more likely to be seropositive to T. gondii. This is likely because this parasite produces tissue cysts that cause lifelong infection and longer lived individuals are more likely to have had opportunities to be exposed to the parasite.2,11
Positive serologic results are not directly indicative of disease status in the individual, nor are they an accurate measure of pathogen impact on population health. If pathogens produce a lasting antibody that is detected by a serologic test, evidence of past exposure may be common in adult age classes, particularly to pathogens that do not cause severe disease. At the population level, seroprevalence for highly virulent pathogens may be exceedingly low or even zero for infections that are almost always fatal. The necessary context for interpreting seroprevalence, which may have an inverse relationship with disease severity, may only be made by accompanying field observations and pathology studies of naturally infected individuals found dead and recovered quickly for a comprehensive necropsy before postmortem autolysis (usually 24 to 72 hours). A large stranding network in California facilitated recovery of beach-cast sea otters for decades, but T. gondii–related disease in otters could be characterized only when a significant investment was made to involve veterinary pathologists who conduct detailed necropsies that included histologic examination of brain tissue. Toxoplasma gondii infection was confirmed in dead stranded otters by immunohistochemistry and culture and isolation of live parasites. Most critically, parasite isolation from infected sea otters enabled mouse bioassay and molecular analysis; this included polymerase chain reaction (PCR) assay and sequencing to confirm the presence of T. gondii and not a related pathogen that might cross-react on serologic assays.1,12,13 Advanced molecular techniques have now differentiated among T. gondii subtypes, or genotypes, infecting sea otters, which may facilitate tracking of pathogen spillover from domestic cat and wild felid sources. Laboratory studies have also greatly facilitated validation of the indirect fluorescent antibody test (IFAT) for use in sea otters. By comparing serologic titers to immunohistochemistry and cell culture results, a cutoff was established that maximized sensitivity and specificity to detect T. gondii infection in sea otters.
Detailed necropsies also enabled characterization of histologic lesions associated with T. gondii cysts in brain tissue so that toxoplasmosis could be evaluated for its role as a primary contributing cause of death in sea otters. Collaboration with facilities that rehabilitate live-stranded sea otters has enhanced interpretation of pathology data by allowing the pathologist to correlate the brain lesions accompanying toxoplasmosis in dead animals with clinical signs observed in those same animals when alive. Once case definition criteria were corroborated, epidemiologic risk factor analyses were used to describe the distribution of T. gondii cases among California otters and evaluate associations between T. gondii infection and more proximate causes of mortality, such as fatal shark-inflicted wounds.7
Armed with a more clearly defined problem of pathogen spillover into this susceptible and threatened wildlife species, research activities have expanded to include field studies of T. gondii distribution and shedding in domestic cats and wild felids in geographic regions with demonstrated high risk for sea otters. Follow-up health investigations in sea otters have developed into long-term longitudinal studies of live animals needed to refine our understanding of the behavioral and ecologic risk factors promoting T. gondii infection in this population. Modern advances in telemetry technology have permitted a direct view into the behavioral complexity of wild animals by improving our ability to observe individual animal movements, foraging activities, contact with domestic animals, habitat usage, and potentially high-risk behaviors. When detailed tracking studies are combined with health and survival indices, these longitudinal studies may identify specific high-risk behaviors that are associated with pathogen exposure. Just as longitudinal studies of people have advanced public health, disease models in sea otters have been used to evaluate a wide array of behavioral traits and movement patterns for their association with T. gondii infection. Modeling efforts have revealed that T. gondii infection in otters is related to the use of specific habitats, which has further refined our assessment of geographic hot spots for pathogen pollution.5 High rates of infection were also identified in sea otters foraging on low-quality prey types, suggesting that increasing rates of protozoal infection in this population may be caused in part by the foraging strategies that have developed to cope with increasingly scarce food resources along coastal California.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree