Chapter 31 The Genus Leptospira
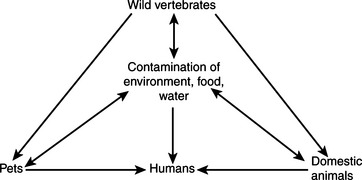
Leptospirosis is encountered commonly by those dealing with animal infectious disease. Its occurrence as a zoonotic infection of humans is strongly influenced by connections to animal production, tropical climates, at-risk occupations, and behavior; it is counted by some as an emerging infectious disease of humans and by others as the most widespread zoonosis. The highest incidence of leptospirosis in the United States is in Hawaii. Interest in human leptospirosis has increased in recent times as a result of several large and well-publicized flood-associated case clusters in Central and South America. This might, as some have suggested, result from increased leptospiral virulence, but it is perhaps more likely to be caused by altered interaction among humans, reservoir species, and the environment.
Until recently, the genus Leptospira was divided into species interrogans (the name deriving from the question mark shape of the organism and comprising pathogenic strains) and biflexa (encom-passing saprophytic, mainly environmental, strains). Growth at 13° C, 8-azaguanine resistance, and failure to form spherical cells in 1 M NaCl distinguished L. biflexa from L. interrogans. Both species were divided into numerous serovars (>60 in L. biflexa and >200 in L. interrogans), and related serovars are placed in serogroups. Many serovars appear to have a certain species as a natural host, but animals and humans can be infected with a wide variety of serovars. The serovars causing disease in animals vary among countries and sometimes among regions in the same country. Most infections in domestic animals are caused by only a few serovars.
Genetic typing of leptospirae has yielded 13 genomospecies (Table 31-1), and this is likely the future of leptospiral taxonomy. However, there is little correlation between serologic type and genomospecies; it is not uncommon for serogroups and even serovars to be represented in multiple genomospecies. Furthermore, methods for genomospeciation are not widely available, and it seems likely that the antigen-based approach to classification will be the standard for some time to come.
DISEASES, EPIDEMIOLOGY, AND PATHOGENESIS
A few key concepts form the foundation for understanding leptospirosis and its pathogenesis. First, leptospires often colonize proximal convoluted kidney tubules and may be excreted in urine for extended periods by reservoir hosts without clinical signs (Figure 31-1). The carrier state may be as short as a few days or may extend throughout the life of the animal. This is considered a major means of host-to-host transmission. Indirect exposure depends on environmental moisture, neutral soil pH, and a sufficiently mild climate to favor survival of leptospires. Streams and ponds contaminated by the urine of wild rodents or domestic and wild animals can be a source of infection for domestic animals and humans, as can urine aerosols in milking parlors (especially those of the herringbone configuration). The organism can also be isolated from milk of infected cows, and this probably serves as a means of transmission to humans and calves (Figure 31-2).
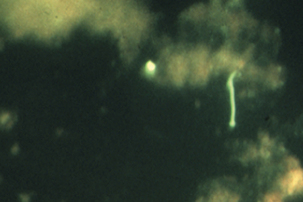
FIGURE 31-1 Fluorescent antibody stain of Leptospira interrogans in kidney.
(Courtesy William A. Ellis.)
Third, many serovars of leptospirae can be roughly categorized as host adapted or nonadapted. Infections by the former tend to be relatively mild and sporadic, with venereal transmission and lifelong colonization of the genitourinary tract; serovars hardjo in cattle and bratislava and tarassovi in swine are examples. Nonadapted strains, however, are more likely to produce catastrophic infections, with abortion storms in pregnant animals and, not infrequently, death of adult hosts. The carrier state is generally brief. Serovar pomona is nonadapted for swine and cattle, as is serovar canicola for dogs. Serovars of serogroups Icterohaemorrhagiae and Ballum are adapted to rats, the latter adapted to mice. The relationship between hosts and adapted strains gives rise to a minor but long-lasting serologic response, whereas nonadapted strains provoke high antibody titers. It should be obvious that a serovar may be adapted to one species and not others. It seems reasonable to speculate that most, if not all, species have host-adapted strains, including perhaps humans.
Finally, there is sufficient leptospiral promiscuity in host selection that it is difficult to say with certainty that a given serovar cannot infect a specific host. Certain strains are more commonly associated than others with disease in a given species, but even that varies with geographic region. Thus it is probably best for veterinary practitioners and diagnosticians to become familiar with the serovars infecting animals in the specific locale in which they practice (Table 31-2).
TABLE 31-2 Examples of Leptospira Serovars Associated with Disease in Specific Hosts
| Serovar | Hosts and Associated Disease |
|---|---|
| L. kennewicki | Equine abortion, repeat breeding (United States, Northern Ireland, England) |
| L. bratislava | Equine abortion, repeat breeding (United States, Northern Ireland, England) |
| Porcine abortion, infertility (worldwide) | |
| L. pomona | Porcine, bovine, equine abortion |
| Skunks | |
| L. canicola | Canine renal, systemic disease |
| Porcine systemic disease, abortion | |
| L. icterohaemorrhagiae | Canine septicemia |
| Bovine, porcine abortion | |
| Classic association with rats | |
| L. hardjo | Bovine abortion, infertility |
| L. grippotyphosa | Canine renal, systemic disease |
| Wildlife infections, especially raccoons, skunks |
< div class='tao-gold-member'>
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree