Reoviridae
Abstract
This chapter describes the properties of reoviruses and features of the diseases they cause in animals.
Keywords
Reovirus; rotavirus; orbivirus; Reoviridae
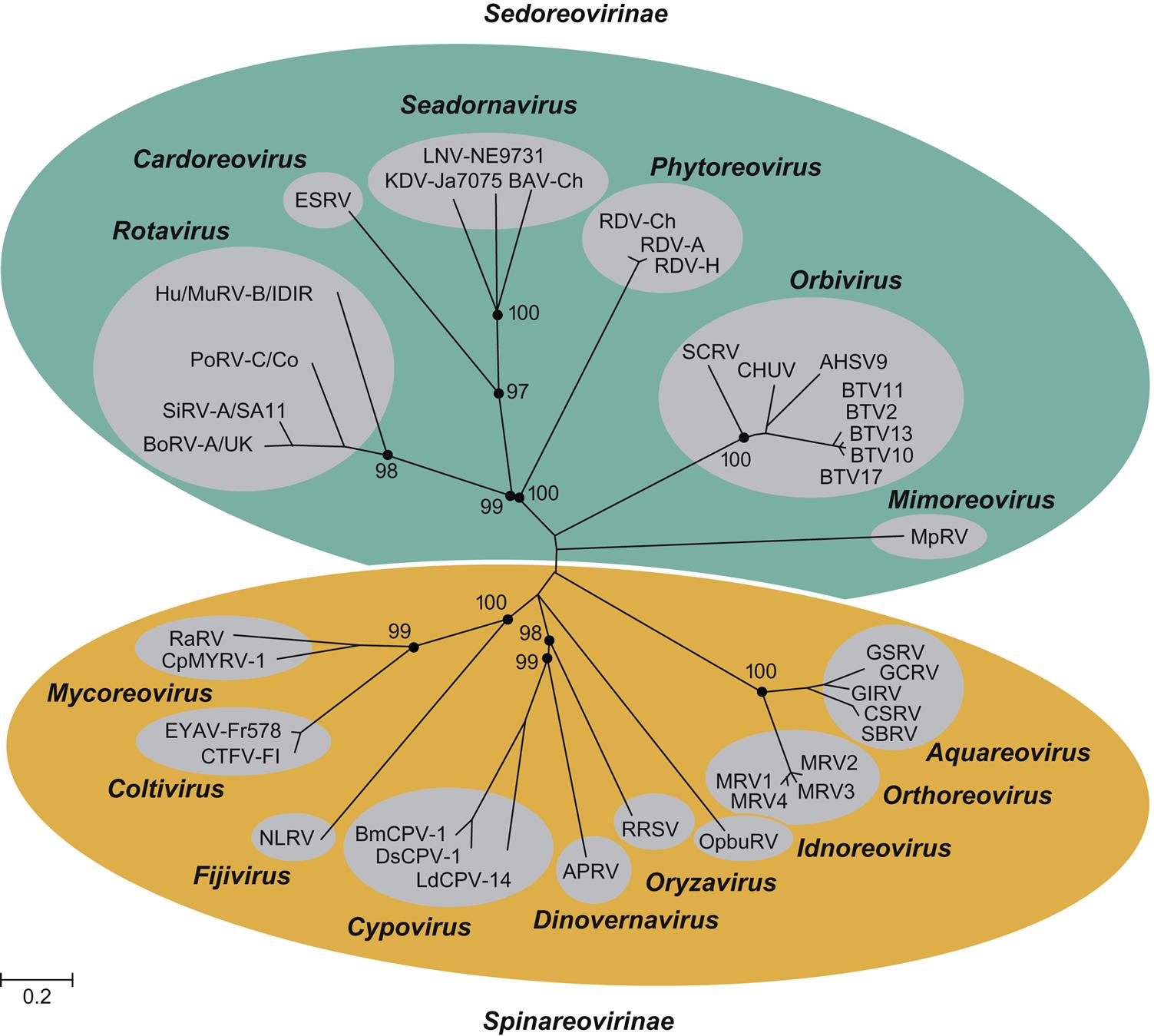
The family Reoviridae is one of the most complex in all of virology, currently comprising 15 recognized genera within two subfamilies of viruses with genomes composed of multiple (10–12) segments of double-stranded RNA (dsRNA) (Fig. 15.1). Individual viruses within the family infect a remarkable variety of hosts, including mammals, birds, reptiles, amphibians, fish, mollusks, crustaceans, insects, plants, and fungi. Viruses in the different genera can be distinguished on the basis of several different features, including their capsid structure, number and size of genome segments, host range and associated diseases, serological properties, and by the nucleotide sequence of their genomes. The root term “reo” is an acronym for “respiratory enteric orphan,” which was coined because the first members of the family were identified in the respiratory and the enteric tracts of animals and humans as “orphans”—that is, not associated with any disease. These viruses are now members of the genus Orthoreovirus. Viruses within the genera Orbivirus, Coltivirus, Rotavirus, Seadornavirus, and Aquareovirus are important pathogens of humans and a variety of animal species, including aquatic animals (Table 15.1).
Table 15.1
Characteristics of Reovirus Infections
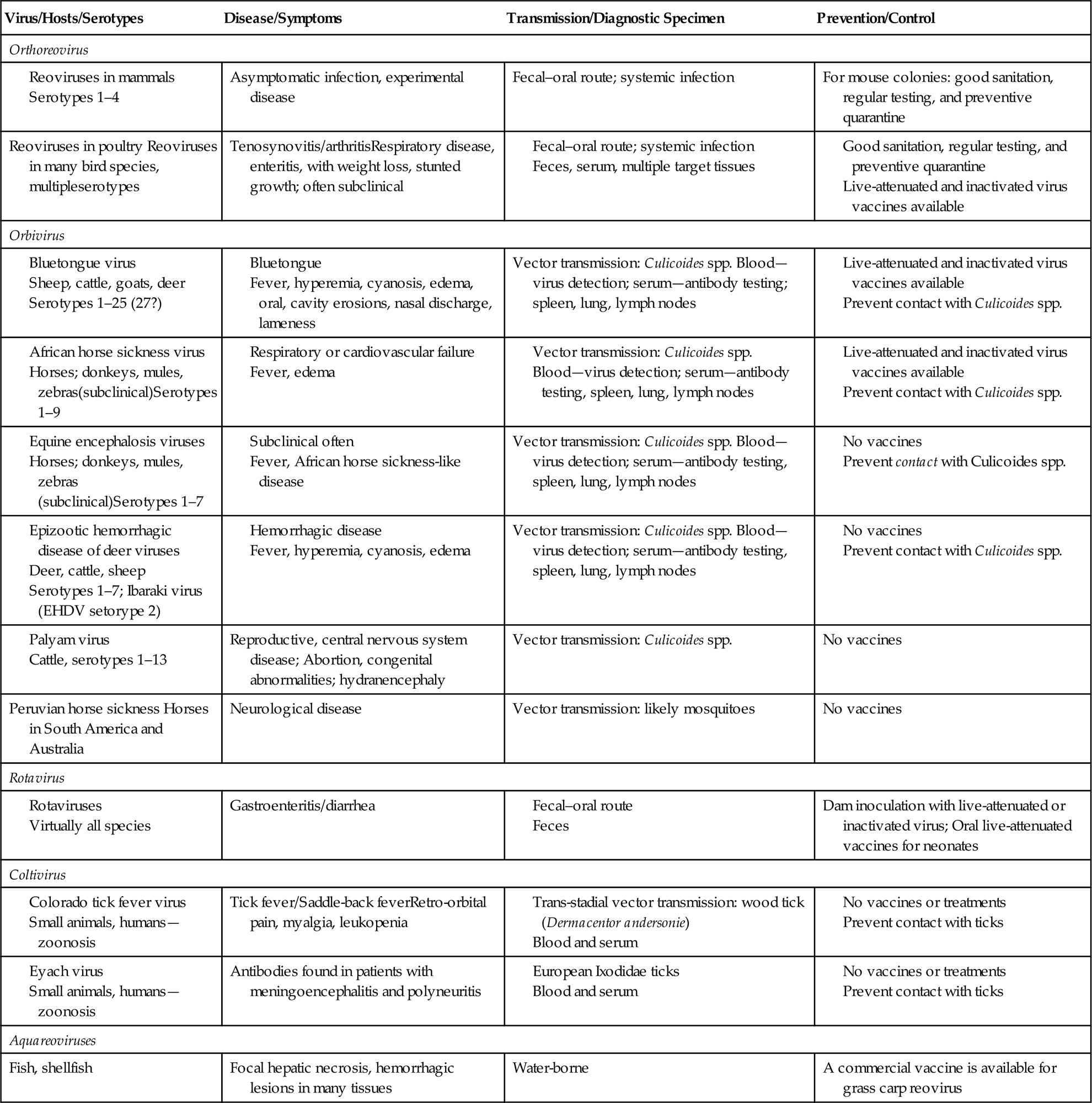
The distribution of the member viruses of the genus Orthoreovirus is ubiquitous, including viruses from cattle, sheep, swine, humans, nonhuman primates, bats, birds, and fish; however, most of these infections are not associated with any significant clinical disease. Only orthoreovirus infections of poultry, and perhaps primates, are of major pathogenic significance. The aquareoviruses most closely resemble orthoreoviruses, both morphologically and phylogenetically, and have been isolated from fish and mollusks in both fresh and sea water. While a large number of aquareoviruses have been described, only a limited number have been shown to serve as primary pathogens.
Viruses of the genus Orbivirus are transmitted to animals primarily by arthropod vectors, which, depending on the individual virus, can be certain species of Culicoides midges, mosquitoes, black flies, sandflies, or ticks. The global and seasonal distribution of individual viruses, therefore, coincides with that of their specific biological vector and appropriate climatic conditions. Bluetongue and African horse sickness viruses are the most important members of this genus from the economic perspective, although several others are potentially important, either regionally or globally. Bluetongue virus infection has been restricted historically to relatively defined areas of the world that include much of the tropical and temperate regions however, multiple different virus serotypes have emerged in the last two decades throughout extensive portions of Europe, perhaps in part as a consequence of climate change in the region. There is, therefore, some concern that African horse sickness and/or other pathogenic orbiviruses might follow.
Rotaviruses are also widespread; essentially, every species of domestic animal and bird has been found to harbor at least one indigenous rotavirus that typically is responsible for causing diarrhea (“scours”) in newborn animals. Rotaviruses cause about 600,000 childhood deaths annually, mostly in developing countries. They also cause considerable economic losses to livestock industries worldwide.
Viruses included in the genera Coltivirus and Seadornavirus are, respectively, transmitted to humans and animals by ticks and mosquitoes. Viruses in these genera are the cause of serious but sporadic human diseases. Genetic analyses suggest a distant evolutionary link between at least some segments or features of coltiviruses and aquareoviruses, and between rotaviruses and seadornaviruses.
Properties of Member Viruses of the Family Reoviridae
Classification
All animal viruses with multisegmented dsRNA genomes are included in the family Reoviridae, with the notable exception of the birnaviruses and picobirnaviruses (see Chapter 16: Birnaviridae and Picobirnaviridae). Consequently, the family is complex, with substantial differences among viruses in the two subfamilies and different genera (Table 15.2; Fig. 15.2). Because of their segmented RNA genomes, reassortment of genome segments among different strains of these viruses is possible within the same virus species, as is a high rate of mutation in individual genes. The resulting genetic shift and drift leads to a remarkable diversity of viruses, which is reflected by the numerous serotypes and myriads of strains of individual viruses within each genus.
Table 15.2
Virions are nonenveloped, spherical in outline, 55–80 nm in diameter
Virions are composed of three concentric capsid layers, all with icosahedral symmetry; the outer capsid differs in appearance in the various genera
Genome is composed of double-stranded RNA, divided into 10–12 segments, total size 18–27 kbp: genus Orthoreovirus, 10 segments, 23 kbp; genus Orbivirus, 10 segments, 18 kbp; genus Rotavirus, 11 segments, 16–21 kbp; genus Coltivirus, 12 segments, 27 kbp; genus Aquareovirus, 11 segments, 15 kbp
Cytoplasmic replication
Genetic reassortment occurs between viruses within each genus or serogroup (genetic shift), along with mutation of individual genes (genetic drift)
Reoviruses are divided into two subfamilies: Spinareovirinae and Sedoreovirinae. The Spinareovirinae comprises nine genera including Orthoreoviruses, Aquareoviruses, and Coltiviruses that infect a variety of animal species. The genus Orthoreovirus comprises at least five different virus species: the mammalian orthoreoviruses, which infect many different species of mammals and include four serotypes (designated MRV1–4); the avian reoviruses including several serotypes isolated from chickens, Muscovy and other species of duck, turkeys, and geese; Nelson Bay orthoreovirus isolated from a fruit bat; baboon reovirus; and reptilian orthoreovirus. The genus Aquareovirus currently contains seven species, designated A–G, that include a large number of viruses isolated from many species of marine and freshwater fish and shellfish. There are only a few members of the genus Coltivirus. Colorado tick fever virus is the prototype virus, and Eyach virus is the European counterpart. A few other coltiviruses have been isolated from other areas of the western United States.
The subfamily Sedoreovirinae comprises six genera that include important pathogens of domestic animals and humans. Within this subfamily are included the genera Orbivirus, Rotavirus, and Seadornavirus. The genus Orbivirus is divided into at least 22 virus subgroups, which represent distinct virus species. Several of these include viruses that cause disease in domestic animals. Separate virus species encompass as many as 27 serotypes of bluetongue virus; 9 serotypes of African horse sickness virus; 7 serotypes of epizootic hemorrhagic disease virus, including Ibaraki virus (a variant of epizootic hemorrhagic disease virus serotype 2); 7 serotypes of equine encephalosis virus; 1 serotype of Peruvian horse sickness virus; 13 serotypes of Palyam virus, including Chuzan virus; certain other viruses affecting different animal species. Individual orbivirus species have been defined both serologically and genotypically, and viruses within each subgroup share a common antigen(s) demonstrable by serologic tests. Genetic differences and relationships among the various viruses can be clearly identified by sequence analyses.
The classification of the member viruses of the genus Rotavirus has historically been based on both genotypic and serologic analyses and currently includes eight species. Rotavirus A includes important pathogens of humans, cattle, swine, horses, gallinaceous poultry, laboratory mice, rabbits, and other animals; Rotavirus B includes pathogenic rotaviruses of humans, cattle, sheep, ferrets, swine, and rats; Rotavirus C includes only pathogens of humans, swine, and cattle; Rotavirus E includes only viruses from swine; Rotaviruses D, F, and G include only pathogens of birds. Historical differentiation of rotaviruses into serotypes is based on neutralization tests. Because both outer capsid proteins (VP4 and VP7) carry type-specific epitopes recognized by neutralizing antibodies, a binary system of classification of serotypes was developed, akin to that used for influenza viruses. For example, in members of Rotavirus A, there are at least 27 [G] genotypes and 14 G serotypes that have been defined on the basis of differences in VP7, and at least 37 [P] genotypes and 14 P serotypes based on differences in VP4. Monoclonal antibodies, gel electrophoresis of viral RNA segments, RNA hybridization, and sequencing of each RNA segment are used to make any further distinctions as may be necessary in molecular epidemiological studies to identify reassortants and potential interspecies transmission events. Sequencing of each genome segment permits a genotype assignment for each gene (specifically, G, P, I, R, C, M, A, N, T, E, H), which further advances understanding of the diversity of each gene segment, its likely host of origin, and overall evolution of rotaviruses.
The genus Seadornavirus includes mosquito-borne viruses that infect humans and animals in Asia, including Banna, Kadipiro, and Liaoning viruses. These viruses initially were included in the genus Coltivirus.
Virion Properties
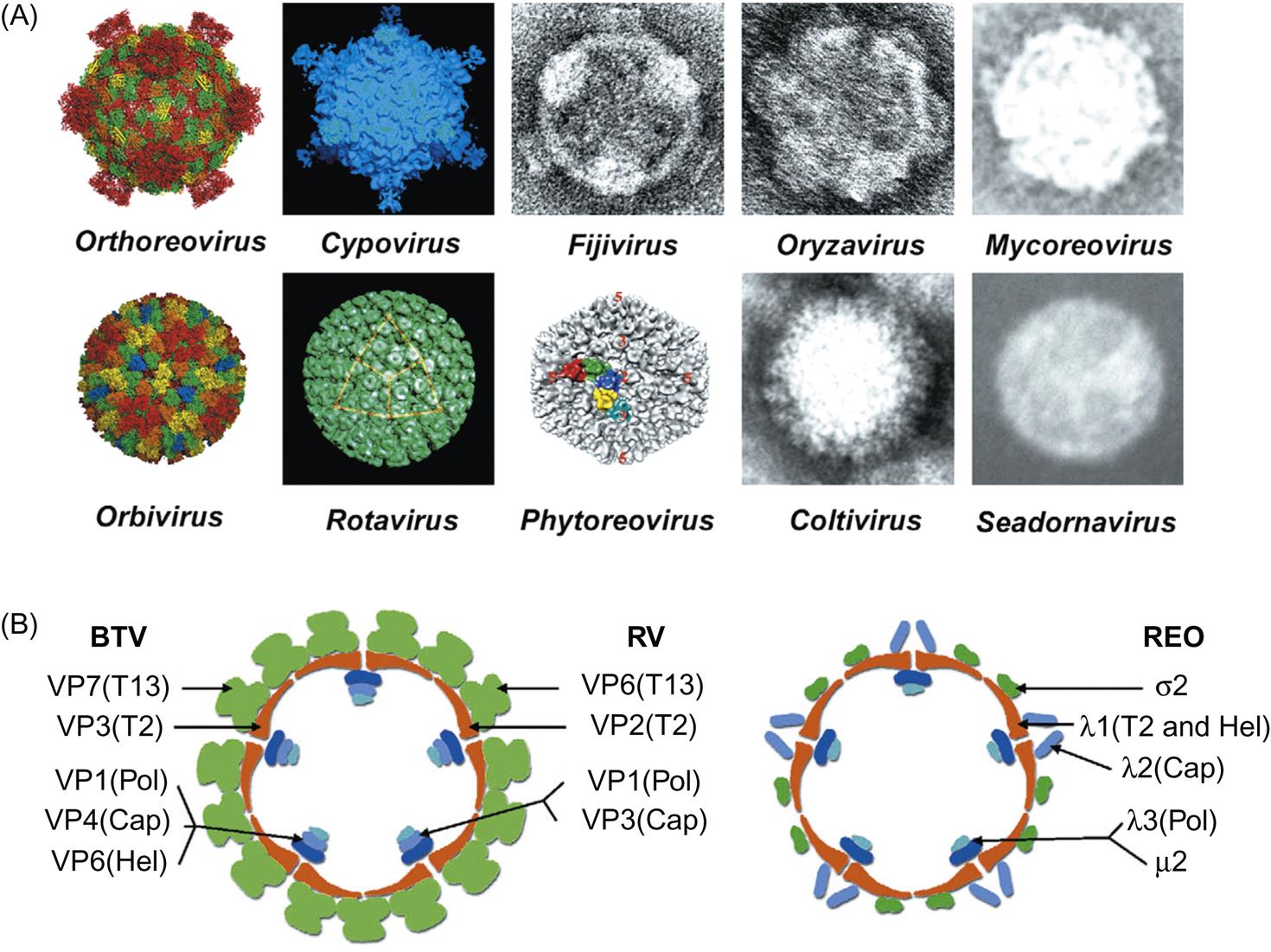
Reovirus particles are nonenveloped, spherical, and have a diameter of approximately 85 nm. Virions consist of a multilayered capsid, each with icosahedral symmetry (Table 15.2). The precise virion morphology varies among the different genera (Fig. 15.2). In general, viruses in the subfamily Spinareovirinae have virions with spikes or turrets that correspond to the 12 icosahedral vertices, whereas viruses within the Sedoreovirinae have a “smooth” appearance. Orthoreoviruses, aquareoviruses, and rotaviruses are resistant to lipid solvents and are stable over a wide range of pH, but orbiviruses and coltiviruses have a narrower zone of pH stability (pH 6–8) and lose some, but not all, infectivity on exposure to lipid solvents. Proteolytic enzymes increase the infectivity of orthoreoviruses, aquareoviruses, and rotaviruses (eg, chymotrypsin in the small intestine cleaves the outer capsid VP4 protein of rotavirus, thereby enhancing infectivity). Proteolytic cleavage of the outer capsid protein VP2 of bluetongue virus (genus Orbivirus) also increases its infectivity for cells of its insect vector (Culicoides species) but not for mammalian cells. Orbiviruses and rotaviruses are remarkably stable. Bluetongue viruses are relatively stable in the presence of protein, and have been reisolated from blood stored for many years at room temperature. Likewise, some rotaviruses are stable for months, even when maintained at room temperature, or for years when stored frozen. Viral infectivity is inactivated by phenols, formalin, 95% ethanol, and β-propriolactone.
The genome of reoviruses consists of linear dsRNA divided into 10 (genera Orthoreovirus and Orbivirus), 11 (genera Rotavirus and Aquareovirus), or 12 (genera Coltivirus and Seadornavirus) segments. The overall genome size is approximately 23 kbp (genus Orthoreovirus), 19 kbp (genus Orbivirus), 16–21 kbp (genus Rotavirus), 29 kbp (genus Coltivirus), 21 kbp (genus Seadornavirus), or 24 kbp (genus Aquareovirus). The positive strands of each dsRNA segment have 5′-terminal caps (type 1 structure). The 3′ termini of both strands lack poly(A) tails. Each RNA segment is present in equimolar proportion in virions. Further detail requires separate mention of each genus.
Genus Orthoreovirus
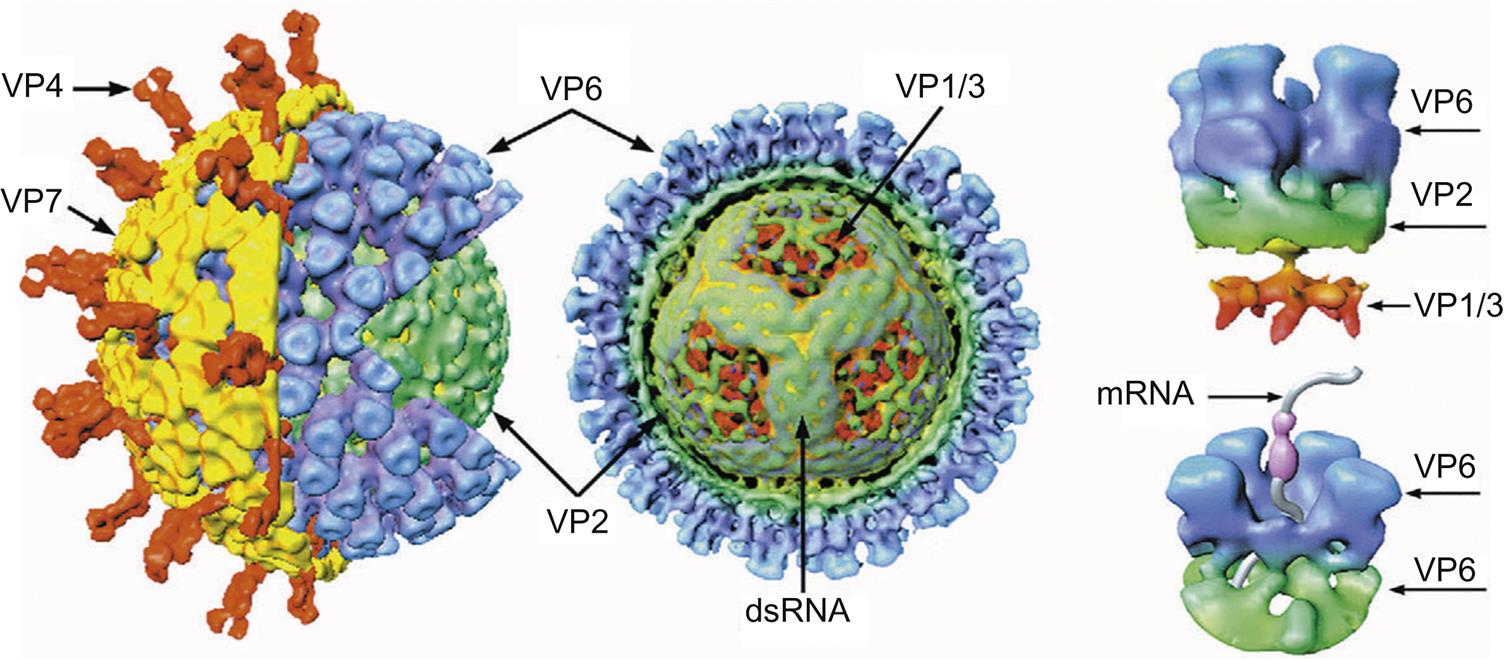
The outer capsid forms a nearly spherical icosahedron, consisting predominantly of complexes of the proteins σ3 and μ1C (Fig. 15.2). In addition to intact virions, there are two stable subviral particles. The first of these is missing only its outer capsid (ie, it is lacking σ3 and contains cleaved forms of μ1 protein); this particle is called the infectious subviral particle (ISVP). The second subviral particle is missing both its outer and its middle capsids and is called the core particle. The protein by which virions attach to host cells, σ1, forms spikes that project through the outer capsid at each of the 12 vertices of the virion. Most importantly, when the outer capsid layer is removed, σ1 protein molecules remain attached to the ISVP and form extended fibers containing the cellular attachment domain at their tips. The core contains the viral RNA polymerase (transcriptase) and consists of three major proteins (λ1, λ2, and σ2) and two minor proteins (λ3 and μ2). The 10 genome segments of the orthoreoviruses fall into three size classes: large, medium, and small. Each segment encodes a single protein, except for one that is cleaved cotranslationally to form two proteins. Each genome segment can be differentiated by size using gel electrophoresis, and these electropherotype patterns have been used to quickly type isolates of mammalian and avian viruses.
Genus Aquareovirus
Virions closely resemble those of the orthoreoviruses morphologically with both outer and inner capsid structures. However, the genome of aquareoviruses consists of 11 segments of linear dsRNA that, like the orthoreoviruses, fall into three size classes that can be differentiated using gel electrophoresis.
Genus Coltivirus
Virions resemble also those of the orthoreoviruses but with a genome consisting of 12 segments.
Genus Orbivirus
The outer capsid consists of a diffuse layer formed by two proteins, VP2 and VP5. This outer capsid is dissociated readily from the core particle, which has a surface composed of 260 trimers of VP7 arranged in ring-like structures for which the genus is named. Both VP2 and VP5 are attached to VP7, which in turn is associated with a subcore shell composed of 120 copies of VP3 surrounding the transcriptase complex (VP1, VP4, and VP6), and the genomic RNA segments. Surface projections are observed only on virions that have been stabilized, otherwise the surface of virions appears smooth and unstructured. The 10 genome segments of orbiviruses encode seven structural proteins (VP1–7) and four nonstructural proteins (NS1–NS4). NS3 is expressed as two isoforms NS3 and NS3A. The genome segments of the orbiviruses are monocistronic, with the exception of the smallest gene segments (9 and 10). Segment 9 encodes a structural and a nonstructural protein in two different coding frames, whereas the single open reading frame of segment 10 encodes a single protein but with two functional initiation codons near the 5′ end of the positive-sense strand. As with the orthoreoviruses, the gene segments have distinct size patterns during electrophoresis that can be used to identify the different orbivirus species. However, with the recent improvement of sequencing technologies, full genome sequencing is now the preferred method to distinguish and compare different virus strains.
Genus Rotavirus
Rotaviruses are triple-layered particles. The outer capsid forms a nearly spherical icosahedron; it consists of the glycoprotein VP7, from which dimers of VP4 extend (Fig. 15.3). The outer capsid of VP7 and the middle capsid, which is composed of VP6 (the structural equivalent of VP7 of orbiviruses), are dissociated readily from the core, which is composed of three proteins: VP1, VP2, and VP3. The 11 genome segments of the rotaviruses are monocistronic, except gene segment 11, which encodes two proteins. In addition to the six structural proteins (VP1–4, VP6, VP7), the rotavirus genome also encodes six nonstructural proteins (NSP1–6). The genome segments can be differentiated by size using gel electrophoresis, and these electropherotype patterns are used to type isolates although full genome sequencing is increasingly applied.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree