Chapter 9 Noninvasive Techniques to Assess Health and Ecology of Wildlife Populations
There are many advantages to noninvasive assessments using samples such as hair, feathers, feces, urine, saliva, regurgitated material, sloughed skin, or even museum specimens. Analyzing urine or feces to evaluate endocrine function without animal capture, restraint, and/or anesthesia minimizes stress and provides a broader measure of endocrine status over a period of hours to days when compared with point in time measures obtained from blood samples. Data collected from fecal hormone and genetic analyses and then overlaid by sample location data may provide information about home ranges, animal movements, stress levels, and habitat use,24 as well as gender and gender ratios. Hair may be analyzed to yield much information about the following: occurrence, distribution, and relative abundance of populations; aspects of population genetics, niche, or diet; detection of rare species; and identification of individuals for wildlife management and forensic purposes.18 Skin samples may be used to create stress response profiles, which may serve as early indicators of health risks.2
Sample Collection and Storage Methods
The tradeoffs and benefits of working with different samples and analytic techniques must be weighed against research objectives and priorities. Frequently, a variety of samples and analytic approaches may be combined to answer research questions in greater depth or provide a broader understanding of wildlife population health (Table 9-1). Through the collection of hair or fecal samples, large remote areas may be surveyed, several population metrics may be calculated, and collection costs are minimal. In some cases, collections must be made noninvasively to avoid sampling bias. The types of samples and collection methods used will vary depending on the species, study objectives, and environmental conditions. For example, hair is collected relatively easily and may be stored for long periods of time but contains smaller amounts of DNA compared with fecal samples. On the other hand, chemical inhibitors present in feces may restrict the amplification of DNA.18
TABLE 9-1 Overview of Biologic Samples, Study Applications, Basic Analytic Techniques, and Collection and Storage Methods*
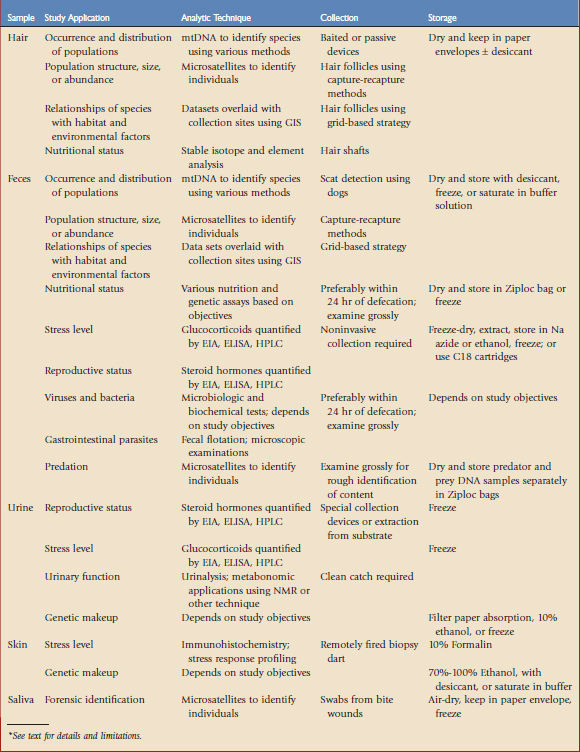
Hair
Determining the occurrence and distribution of populations is a common goal of hair collection studies, and hair may be used to answer questions about population genetics and structure when DNA quality is high. However, estimating population size or abundance via hair collection surveys is not as effective because it depends on reliable individual identification based on nuclear DNA analysis, and collection methods may not be efficient enough to provide a sufficient capture-recapture sample size. Population trends may be estimated by repeatedly monitoring occupancy if detection probability may be determined, and these data may guide wildlife management decisions (e.g., documenting wildlife use of highway crossing structures). Evaluating relationships of species with habitat and human variables using grid-based sampling, combined with identification of individuals through genotyping, is possible,1 and hair samples may be analyzed in nutritional studies as well.
The best source of DNA is follicles, which are more frequently present on plucked as compared with shed hair,10 although hair shafts may provide useful DNA contributed by saliva, dander, or other adherent tissue.26 Hair should be collected within 3 to 4 weeks of deposit for the best genotyping results because ultraviolet light and moisture degrade DNA over time. Hair may be collected opportunistically or with sampling devices, which may be passive or baited. Passive devices collect hair during normal behavior and include hair-snagging devices mounted on natural rub objects (e.g., on trees for bears) or along travel routes (e.g., barbed wire strands for badgers). Baited sampling devices include food or scent lures to attract animals to collection sites. Catch structures should be designed to minimize collection of hair from multiple individuals or species, and variables such as animal behavior, movement, concentrations, and collection intervals should contribute to design strategies. Hair should be stored dried in small paper envelopes or vials with silica gel desiccant until it is analyzed.
Feces
A single fecal sample may provide information about reproductive status, genetic makeup, stress, viruses, internal parasites, predation, and diet. Noninvasive capture-recapture survey techniques may be adapted to estimate population size by supplementing detection data with genetic analyses of hair or scat samples, indigestible plastic chips recovered in scat, or cameras to identify individuals.20 Capture-recapture methods through scat collection may be successful if the target population is not of extremely low density, sampling methods have a high rate of detection and low level of sampling bias, population size does not change between collection periods, and accurate identification of individuals is possible.14 Collection of fecal samples may be greatly facilitated by the use of dogs that have been trained specifically to search for wildlife scats.24 Dogs may even be trained to distinguish individual animals by their scat. However, detection rates may vary among dog-handler teams, making proper training and a study design that allows for estimation and correction of detectability highly important. Thorough mixing of a fecal sample prior to collecting a subsample will help ensure reliable results because corticosteroids and their metabolites may be unevenly distributed in feces.15
There are several ways that fecal samples may be stored, depending on the analyses to be performed. For endocrine assessment, sodium azide or other preservatives such as ethanol may be mixed with feces to prevent bacterial growth because bacteria and their enzymes will degrade steroid metabolites in a few hours. Alternatively, fecal samples may be stored frozen at –20° C for preferably no longer than 90 to 120 days prior to extraction and analysis.13 A field-based extraction and storage technique using C18 cartridges (Varian, Walnut Creek, Calif) makes it possible to store samples at ambient temperatures for up to 2 weeks before freezing or analysis.3
For nutritional studies, fecal samples should ideally be collected within 24 hours of defecation, and indicators may include degree of wetness and lack of insect damage. Dietary components may be determined by gross physical examination of fecal samples as well as laboratory analysis to determine diet preferences of various species, assess nutrient content, and monitor forage quality. Molecular tools have forensic applications as well—for example, in areas in which predation on livestock is suspected.8 For carnivore species, scats may be air-dried on sterile paper, prey remains examined grossly for rough identification and subsequent DNA analysis, and bile powder separated to identify predator species. Dried samples should be stored in Ziploc bags in a dark, dry location until DNA extraction. Fecal samples from herbivores should be dried at 80° C for 48 hours, ground into a fine powder, and then stored frozen until time of analysis.
Urine
Urine samples, like fecal samples, may be used for endocrine evaluations and genetic analyses.12 Urinary DNA is easier to extract and analyze than fecal DNA; however, potential gender-specific differences in urination behavior may affect estimation of population size and gender ratios. Urine samples may be collected from captive wildlife housed in exhibits with concrete flooring and may also be extracted from natural substrates such as snow or sand.16 Simple collection devices may also be devised (e.g., containers mounted on the end of sticks to catch urine from arboreal primates). Although collection of urine samples in the field may occasionally be challenging compared with fecal samples, urine requires no further processing before being assayed in the laboratory and may be preserved by absorption onto filter paper, mixing with 10% ethanol for storage at room temperature for up to 12 weeks, or freezing indefinitely.18
Saliva
Saliva has been used for determination of blood glucose and plasma steroid hormone concentrations, as well as detection of several diseases. Salivary concentrations of testosterone, 17β-estradiol, and cortisol correlate best with blood levels 20 to 40 minutes prior to collection in humans and sheep. Samples of saliva may be collected using a bulb, which requires some animal handling but, compared with venipuncture, these minimally invasive techniques are less stressful. Salivary DNA may be recovered from carcass bite wounds to identify the species, gender, and individual identity of the predator.26 For genetic testing, swab samples should be air-dried for 24 hours, sealed in a paper envelope, and stored in a plastic bag at –20° C until they are analyzed.
Supplementary Observational Techniques
Behavioral observations, track surveys, images from remote cameras, and movements monitored via radio or satellite telemetry units may augment data acquired from field samples and help validate assays. Surveys of tracks in mud or dust may assess the presence and distribution of animals in remote or relatively inaccessible areas (e.g., aerial track survey), but determination of relative abundance requires identification of individual animals. Natural sign surveys have been documented for numerous animals and may be affected by adjacent habitat characteristics and environmental conditions. Population size may be estimated when the probability of scat detection is known; this depends on an understanding of the rates of scat decomposition and defecation,14 especially when search areas cannot be cleared of scat (as by a snowstorm) prior to surveying.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


