Chapter 83 Mycoplasma haemolamae in New World Camelids
In 1990, the first descriptions of a newly identified organism found on red blood cells of llamas were published in two reports from Colorado and Kentucky.6,10 The organism was called an Eperythrozoon-like organism, based on light and scanning electronic microscopic morphology.
In the initial reports of infections with this organism, some affected llamas had mild to severe anemia that was associated with the variable presence of anisocytosis, reticulocytes, and nucleated red blood cells. Some of the more severely anemic llamas also had low serum iron levels, hypoalbuminemia, and hypoglycemia. These severely affected llamas also tended to have poor body condition and evidence of other concurrent disease. Other infected llamas did not have significant clinical signs nor hematologic or biochemical abnormalities.6,10
Sera from some of these infected llamas reacted positively with antigen from Eperythrozoon suis–infected swine red blood cells (RBCs), leading to speculation that this organism could be a previously named Eperythrozoon or Hemobartonella that had found a new host species.6,7 Attempted transmission to cats, sheep, and swine was unsuccessful, even when the cats and pigs were splenectomized. This suggested that the camelid organism was, in fact, a new species.
Johnson and colleagues5 attempted to reproduce the infection in llamas experimentally by intravenous or subcutaneous injection of infected blood into six healthy adult and yearling llamas. Only one of these llamas became more than transiently infected, as detected by examination of blood smears. However, later dexamethasone suppression showed that some of these llamas did appear to be infected. This early experiment suggested both that infectivity of the organism might be relatively low in healthy animals and that a chronic carrier state might exist.
Reports in 1992 and 1997 of eperythrozoonosis in immunodeficient llamas provided support for the idea that the organism caused clinical disease, primarily in camelids that were not immunocompetent.2,4
Polymerase Chain Reaction Assay Development
In 2001, a polymerase chain reaction (PCR)–based assay was developed to provide a more sensitive and specific diagnostic technique for diagnosis of the infection and to study the kinetics of infection, response to treatment, and prevalence of infection.11,12 The assay was developed using blood from a naturally infected alpaca from Oregon. The primers for the assay were chosen to amplify a 318–base pair (bp) sequence that was unique to the hypervariable region of the organism’s 16S rRNA region, as identified in GenBank accession AF306346.8 Specificity of the assay was demonstrated by failure of the primers to amplify several related organisms. The lower detection limit was shown to be approximately 28 gene copies, or one organism in 3.8 to 7.7 × 109 erythrocytes. The assay was able to detect the organism in samples from llamas and alpacas from almost every state in the United States, as well as a variety of locations in Canada, Australia, and the United Kingdom, thus demonstrating that the assay was not geographically limited.
Classification and Characteristics
In 2002, Messick and associates9 reported that the 16S rRNA sequence of an Eperythrozoon-like organism amplified from a heavily infected alpaca was closely related to the hemotropic mycoplasmas present in cattle, pigs, cats, and opossums. The new name, Candidatus Mycoplasma haemolamae was proposed for this organism based on morphologic, genomic, and structural characteristics. Although this organism has not yet been cultured, precluding an official species designation, it is often referred to as Mycoplasma haemolamae.
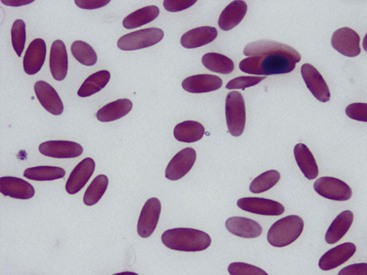
M. haemolamae, along with the other related hemotropic mycoplasmas, are small (0.4 to 0.6 µm) bacteria that lack a cell wall. They may appear as coccoid, linear, or ring-shaped and most often appear to be on the edge of the erythrocyte or free in the background, especially if there is a delay between blood collection and making blood smears (Figs. 83-1 and 83-2). Scanning electron micrographs have shown that there is often a slight depression in the red blood cell membrane where the organisms are located, but the bacteria do not actually enter the cell.10
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree