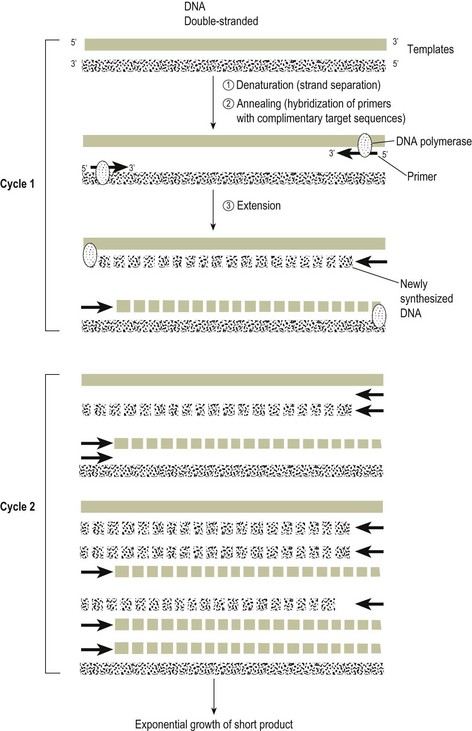
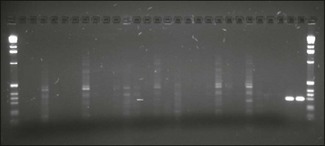
Chapter 4 A diagrammatic representation of the basic PCR procedure is shown in Figure 4.1. PCR is a cyclical process of copying DNA, which involves heating and cooling. Each cycle has three steps, the first of which involves the denaturation of double stranded DNA. The DNA double helix comprises two phosphate sugar (deoxyribose) polymers that are connected by hydrogen bonding between the bases adenine (A), guanine (G), thymine (T) and cytosine (C). Guanine pairs only with cytosine and adenine with thymine. The PCR method separates the two strands of the DNA helix by heating at a temperature of 93 to 94°C. The hydrogen bonds between the paired bases are disrupted giving rise to single stranded DNA. Figure 4.1 Diagram of PCR reaction. The products generated at cycle 1 are not of a defined length. The first PCR products of defined length are produced at cycle 2 due to priming on the products generated during cycle 1. A third cycle results in the first double-strand PCR products of defined length and subsequent cycles lead to exponential amplification of PCR product. The PCR cycle of denaturation, annealing and extension is repeated and the product resulting from the extension of one primer serves as the template for the second primer and vice versa. Initially synthesis goes beyond the sequence complimentary to the other primer but with each cycle of denaturation, annealing and primer extension the target, that is, the region of DNA flanked by the primers, increases almost exponentially. The PCR cycle is repeated 30 to 40 times to produce detectable amounts of PCR product or amplicon. The simplest way to detect the PCR product is to load a fraction of the reaction onto an agarose gel (Fig. 4.2). The product should be visible as a sharp band of expected size. Fluorimeter-based real-time detection systems (see below) are replacing conventional PCR and gel-based detection systems in many veterinary laboratories. Zinc-finger proteins, DNA-binding proteins that directly and specifically detect PCR products, have recently been used to detect microorganisms including Salmonella spp. and influenza A viruses. Figure 4.2 PCR products, following amplification of signature sequence of 16S region of chlamydiae, electrophoresed in agarose gel and stained with ethidium bromide. Two lanes (far right) contain amplicon of correct product of size 298 base pairs. First and last lanes contain a molecular size marker.
Molecular techniques in diagnostic microbiology
Polymerase Chain Reaction (PCR)
< div class='tao-gold-member'>
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Molecular techniques in diagnostic microbiology
Only gold members can continue reading. Log In or Register to continue
