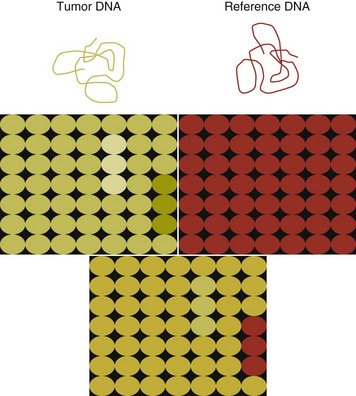
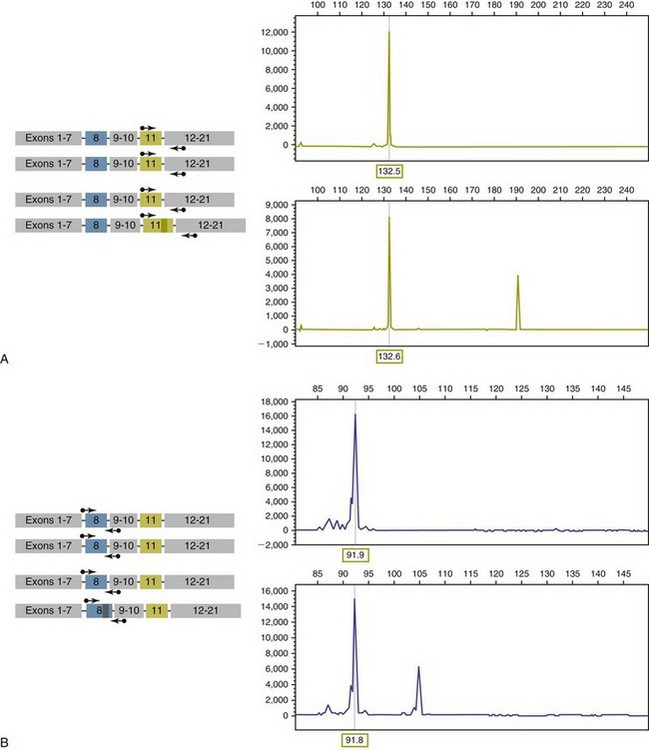
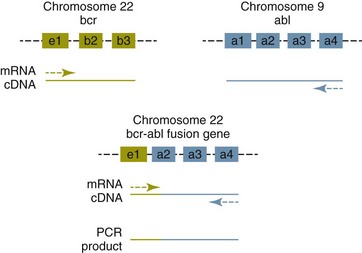
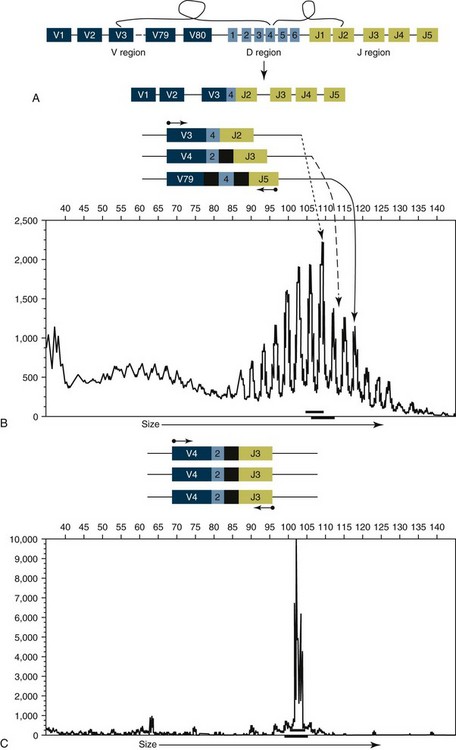
8 Since the mid-1980s, advances in the fields of molecular biology and genetics have changed our understanding of the biology of cancer. Technologic advances now provide the opportunity for applying this advanced understanding to the clinical arena in the form of novel diagnostic tests and management strategies. Molecular approaches for cancer diagnosis are now part of the standard of care for most human patients.1 Technologic advances have reduced the unit cost for many of these approaches, and their use and commercialization is increasingly taking hold within the veterinary field. It is now feasible to assess proteins, RNA, DNA, or their metabolites in tumors, effusions, blood, saliva, and urine to more accurately classify and detect various forms of neoplasia. Recently, the National Comprehensive Cancer Network (NCCN) defined molecular testing in oncology as “procedures designed to detect somatic or germline mutations in DNA and changes in gene or protein expression that could impact the diagnosis, prognosis, prediction and evaluation of therapy for patients with cancer.”1 Molecular assays are used for a variety of reasons in the diagnostic evaluation of malignancy. For example, the presence or absence of individual oncogene mutations or chromosomal abnormalities has significant prognostic implications in many types of malignancies. In people, finding a mutation in the nucleophosmin gene in cases of acute myelogenous leukemia (AML) predicts a significantly better prognosis, independent of other factors.2 In canine mast cell disease, the presence of a mutation in the c-kit gene suggests a worse prognosis than for patients without such a mutation.3 In lymphoma, the phenotype (B- versus T-cell) plays a large role in determining prognosis in both veterinary and human patients. A variety of molecular diagnostic tests, including DNA-based and various methods of protein detection, can establish the phenotype of lymphoma. The presence of a particular mutation or chromosomal abnormality can help to subclassify a tumor. For example, chronic lymphocytic leukemia/small cell lymphoma (CLL/SCL) and mantle cell lymphoma in humans are both neoplasms of mature B-cells, with a similar (but not identical) immunophenotype. Mantle cell lymphoma, however, almost always has a rearrangement between the immunoglobulin heavy chain locus (IgH) and the CCND1 (encoding cyclinD1) gene, whereas this rearrangement is very rare in CLL/SCL.4 The prognosis and treatment of these two diseases is quite different, so the distinction is important to make. Molecular diagnostic testing also helps guide therapy. This may be best illustrated by the development of tyrosine kinase inhibitors (TKIs). These drugs inhibit signaling through tyrosine kinase receptors, such as c-kit, platelet-derived growth factor (PDGF) receptor, and epidermal growth factor (EGF) receptor. Tumors with mutations in these receptors that result in their constitutive activation respond well to TKIs, whereas those without may require different kinds of chemotherapy. Thus testing for mutations in these genes has become commonplace in human medicine—EGF receptor in small cell lung carcinoma and stem cell factor (SCF) receptor (c-kit) in gastrointestinal stromal cell tumors. Similarly, mast cell tumors in dogs that harbor a c-kit mutation respond better to TKIs than those without the mutation.3 Finally, oncogenes and chromosomal translocations uniquely distinguish neoplastic from normal tissue. As such, sensitive detection of mutations can be used to quantify residual disease in patients that have been treated. The best example of this is detection of the bcr-abl fusion gene, which can allow oncologists to detect as few as 1 : 106 neoplastic cells in peripheral blood.5 Tumor-specific primers that recognize the unique immunoglobulin genes found in both canine and human B-cell lymphomas have been used to quantify tumor burden and monitor disease in both dogs and people with lymphoma. Historically, these techniques involved the examination of metaphase preparations made from chromosomes. Metaphase preparations were then stained (banded) to help in the identification of distinct chromosome morphologies. Using these techniques, detection of gross abnormalities in chromosome number (ploidy) and of the presence of chromosomal translocations was possible and led to the identification of genes associated with tumor development and progression. Cytogenetic analysis has been most useful in the clinical assessment of leukemias, in which metaphase preparations are relatively easy to develop from whole blood samples.6 For most human leukemias, cytogenetic descriptors are used to define distinct subgroups into prognostic groups and to guide treatment decisions. The use of cytogenetic approaches in the management of companion animals has been limited due to the difficulty in using conventional chromosomal banding to identify canine chromosomes. The development of chromosome-specific “paints” that allow the identification of specific canine chromosomes has improved the opportunity to apply cytogenetic descriptors to canine cancers. Using these techniques, Breen and colleagues have identified a chromosomal translocation in canine chronic myelogenous leukemia (CML) and chronic monocytic leukemia that is the equivalent of the bcr-abl Philadelphia chromosome found in human CML.7,8 For the most part, traditional cytogenetic techniques, including the use of chromosome-specific paints, are labor intensive and have been replaced by alternative modalities. Comparative genomic hybridization (CGH) arrays can define gains and losses in chromosome number within tumor specimens rapidly and with highly reproducible results. In CGH analysis, the investigator labels genomic DNA from a normal individual and from tumor cells of the patient with two different color fluorescent probes. The labeled DNA is then hybridized to an array of DNA probes that span the majority of the genome. These probes are printed onto a chip or slide, such that the location of each individual probe is identified. The degree of hybridization to each probe is then determined by the level of fluorescence detected by laser excitation. Equal hybridization of the DNA from both sources to an individual probe indicates normal copy number, whereas increased binding by the tumor DNA indicates the presence of chromosomal duplication in the area of the genome covered by that probe (Figure 8-1). Similarly, higher binding by the DNA from the normal individual indicates chromosomal loss in the area. CGH arrays are useful for localizing chromosomal regions where investigators should focus their search for genes important to that cancer. A canine CGH array with 2 megabase resolution has been reported.9 Using this array, Breen and colleagues have shown that a subset of T-cell lymphomas (histologically defined as peripheral T-cell lymphoma, unspecified) exhibits copy number gain in regions common to most examples of this histologic type but not present in other T-cell lymphoma subtypes.10 This finding will help to identify genes within the duplicated areas that might be useful for diagnostics and therapy and for understanding the genesis of the neoplasm. A similar study in canine malignant histiocytosis (MH) demonstrated that MH frequently exhibits loss of chromosomal regions that contain tumor suppressor genes.11 Such genetic characterization of a diverse group of cancers, with distinct biologic behaviors but similar histologic descriptions, will significantly improve opportunities to target specific therapies and management strategies to distinct biologic subgroups of these diseases. Primer synthesis typically requires that the sequence of the target gene is known. The publication of the canine genome in 200512 has been invaluable in this regard, since it is now possible to simply use the known canine sequences rather than hope for sequence similarities with mice and humans. PCR-based assays are commonly used in human oncology to detect insertions or deletions in genes relevant to the prognosis or treatment of a neoplasm. In veterinary medicine, detection of internal tandem duplications (ITD) in the c-kit gene in canine mast cell tumors is now a routine part of the diagnosis for the purpose of determining the most effective treatment. C-kit is a TK receptor for the growth factor SCF and is mutated in some cases of canine mast cell tumors.13 The primary mutations described are internal ITD in two different exons, exon 8 and 11.14 The mutations involve the duplication of a small segment of DNA so that it is repeated, resulting in a larger gene (Figure 8-2). Approximately 14% to 20% of canine mast cell tumors have a duplication in either exon 8 or 11.14 Since mast cell tumors containing the ITD respond better to TK inhibitors than tumors with wild-type c-kit,15 testing for c-kit mutations has become a routine part of the diagnostic work-up for mast cell tumors. Detection of this type is fairly simple, since the presence of a larger (or smaller, in the case of deletions) PCR product is determined by size separation. It is likely that as more genes are identified as being targets of therapy, such assays will become more frequent. Recently, Suter et al identified an internal duplication in the FLT3 gene16 in acute lymphocytic leukemia (ALL) using the same methods and provided preliminary evidence that response to a small molecule inhibitor is predicted by the presence of the mutation in lymphoid cell lines. We are likely to see routine use of mutation detection grow in the near future and continue to guide treatment decisions. One mechanism by which chromosomal translocation causes malignant transformation of cells is to create novel proteins with altered function. The best studied of these fusion genes, the Philadelphia chromosome, is the breakpoint cluster region-Abelson (bcr-abl) fusion gene found in greater than 90% of all human CMLs and occasionally ALL and AML.17 abl is a tyrosine kinase that has myriad activities involved in cell growth and differentiation. It is encoded on human chromosome 9, and in CML, it is translocated to chromosome 22. The site of the translocation varies within the bcr gene, so that a new fusion gene, bcr-abl, is formed. The new fusion protein allows for the constitutive activation of the abl tyrosine kinase, which in turn promotes the development of CML (Figure 8-3). This fusion protein is the product of a novel RNA transcript, which can be readily detected by PCR. To accomplish this, RNA is extracted from blood containing potentially neoplastic cells and reverse transcribed to cDNA. The cDNA is then amplified with two primers, one which anneals to the bcr gene, and the other to the abl gene. Since these two genes are normally on different chromosomes, no product will be seen in the absence of a translocation, but will be detected if the genes have been brought together to form a single messenger RNA (mRNA). This assay can detect as few as 1 : 106 tumor cells5 and can therefore be used for both diagnosis of CML and quantifying residual disease after treatment. Assays for a large number of translocations have been developed over the past 10 years.18 These assays are now routinely available for characterization of human tumors, particularly leukemia. The finding that canine leukemia and lymphoma can exhibit the same translocations as their human counterparts8 suggests that detection of this novel fusion gene would aid in the diagnosis of canine chronic myelogenous leukemia. A clonality assay demonstrates that a group of cells is derived from a single clone. The term is usually used to refer to detection of the unique genes found in each individual B- or T-cell—immunoglobulin genes in B-cells and T-cell receptor (TCR) genes in T-cells. The portion of these genes that encodes the antigen-binding region is the portion that varies between cells, both in size and sequence. When B- or T-cells divide, the immunoglobulin and TCR genes are passed on to the daughter cells.19,20 In the course of a normal immune response to a pathogen, B- and T-cells are activated, expand, and eventually die, leaving behind a small number of residual memory cells. On the other hand, when a cell becomes neoplastic, it no longer responds to growth controls and undergoes unlimited expansion. Therefore, if one can establish that the majority of cells in a particular collection of lymphocytes have the same immunoglobulin or TCR gene, it is most likely that these cells are neoplastic rather than reactive.21 When immunoglobulin and TCR genes rearrange during the course of B-cell and T-cell development, respectively, the length and sequence of the resultant gene differs from cell to cell. There are many reasons for this, including the fact that nucleotides are added between V, D, and J segments as they rearrange into a contiguous formation. The clonality assay takes advantage of this fact. In a sample consisting of many different lymphocytes, as in a reactive process (the lymph nodes of a dog with chronic pyoderma or poor dental hygiene, for example), there will be multiple different-sized TCR and immunoglobulin genes. On the other hand, in a sample consisting of neoplastic lymphocytes, the immunoglobulin gene or the TCR gene (depending on whether it is a B-cell or a T-cell lymphoma) will be a single size (Figure 8-4). Clonality assays are accomplished by isolating DNA from cells suspected to be neoplastic and then using PCR primers directed at the conserved regions of TCR or immunoglobulin genes. The primers amplifying the variable regions and the PCR products are separated by size using a variety of possible methods. The presence of a single-sized PCR product is indicative of clonality, whereas the presence of multiple PCR products supports a reactive process. This assay has now been reported by a number of laboratories22–25 and used to answer a variety of clinical questions.26–28 This assay is termed the PCR for antigen receptor rearrangement (PARR) assay in order to distinguish it from other types of clonality assays.23 It should be noted, however, that the term PARR is not used in the human literature, where the assay instead is referred to as a clonality assay. The principle of the clonality assay can also be used to quantify tumor cells in blood or node and to monitor minimum residual disease. For this type of analysis, PCR primers specific for the immunoglobulin or TCR gene that is carried by the tumor are used, instead of the broadly reactive primers used to screen samples. In this way the investigator is certain that only tumor DNA is being amplified and not nonneoplastic lymphocytes. The specificity of this reaction permits determination of the number of tumor cells in a sample of blood, even when those cells are as rare as 1 : 10,000 cells. Yamazaki et al28 demonstrated that with current chemotherapy protocols, all seven dogs they examined had at least 1 : 10,000 cells in their peripheral blood, even though the dogs achieved clinical remission. Although this kind of analysis may not be practical for routine diagnostics, it is a powerful research tool to compare the efficacy of novel chemotherapy regimens.
Molecular Diagnostics
Goals of Molecular Diagnostic Testing in Oncology
Methods for Analyzing Genes
Detection of Chromosomal Abnormalities
Polymerase Chain Reaction–Based Techniques: Detection of Mutations, Novel Genes, and Assessment of Clonality
Detection of Genetic Insertions and Deletions
Detection of Fusion Gene Products by PCR
Assessment of Clonality in Lymphoma and Leukemia
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Molecular Diagnostics