Circoviridae and Anelloviridae
Abstract
This chapter describes the properties of circoviruses and anelloviruses, and features of the diseases they cause in animals.
Keywords
Circovirus; Gyrovirus; Circoviridae; Anelloviridae; Parvoviridae
Although circoviruses and anelloviruses are biologically and taxonomically distinct, viruses in the two families are morphologically similar and have genomes of circular single-stranded DNA. Together with viruses in the family Parvoviridae, members of the families Circoviridae and Anelloviridae are the smallest known DNA viruses of vertebrates. The family Circoviridae includes important pathogens of birds and swine, and perhaps dogs. The family Anelloviridae includes an increasingly extensive collection of genetically diverse viruses that infect a wide variety of animal species, notably humans and swine, although their clinical significance is currently uncertain.
Members of the Family Circoviridae
Properties of CIRCOVIRUSES
Classification
The member viruses of the family Circoviridae share some common virion and genome properties, but are ecologically, biologically, and antigenically quite distinct. Circoviruses are similar to plant geminiviruses and nanoviruses in terms of their genomic organization and replication strategy. It is speculated, therefore, that animal circoviruses may have originated from a plant nanovirus through host-switching and subsequent recombination with a mammalian virus. The family currently includes two genera (Circovirus, Gyrovirus). Porcine circovirus 1 is the type species of the genus Circovirus, members of which use an ambisense genome replication strategy with viral genes in different orientations. The genus Circovirus includes beak and feather disease virus, canary circovirus, goose circovirus, pigeon circovirus, duck circovirus, finch circovirus, gull circovirus, and porcine circoviruses 1 and 2. A recently described canine circovirus shares the same general properties of these circoviruses, as do many other avian circoviruses that are not yet classified taxonomically. Chicken anemia virus is the type member of the genus Gyrovirus, in which the viral genes are all in the same orientation. Additional genetically diverse gyroviruses have recently been identified in humans and chickens, although the biological and clinical significance of these novel gyroviruses is uncertain. The “Cyclovirus” sequences recently detected in feces from humans and chimpanzees may form an additional, previously unrecognized genus in the family Circoviridae.
Virion Properties
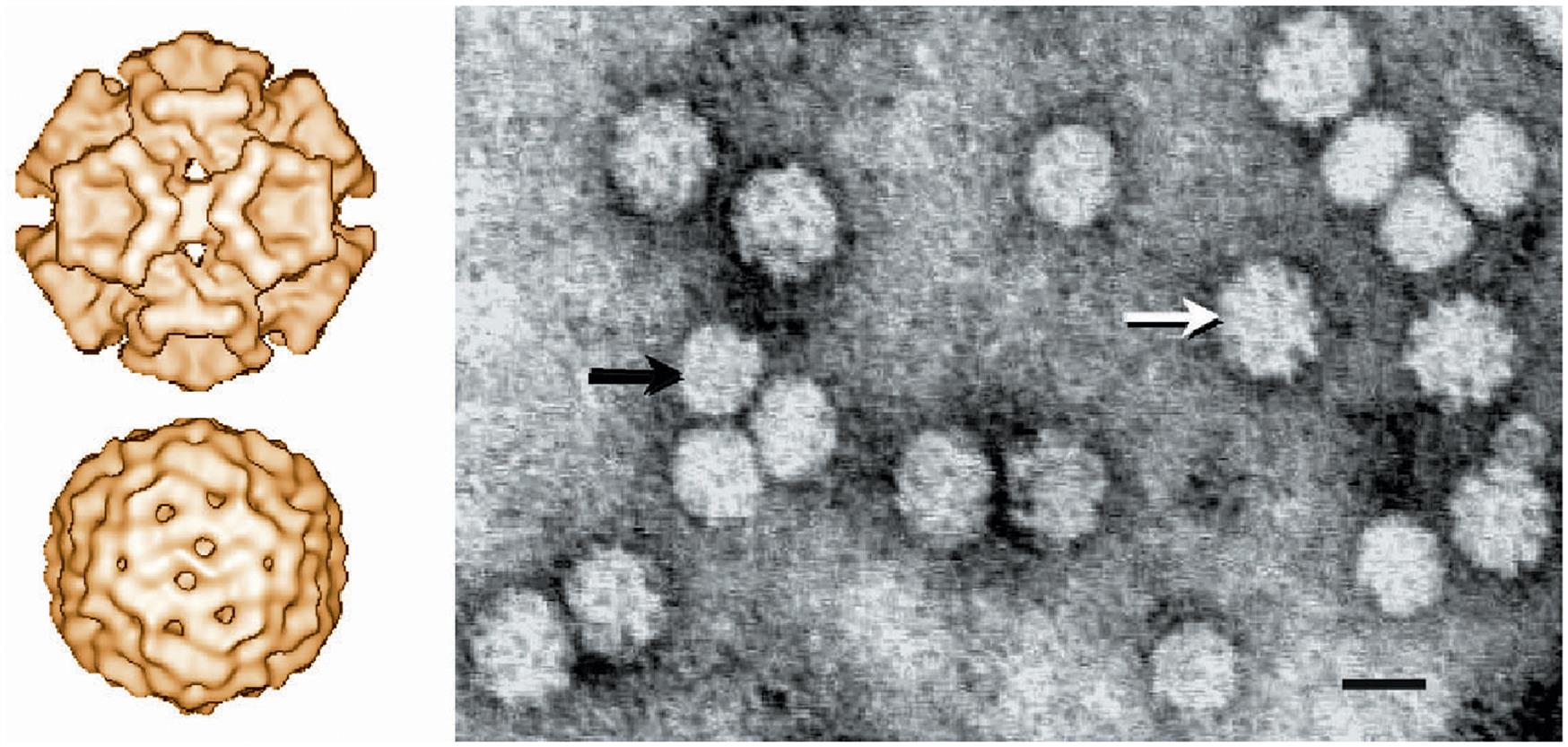
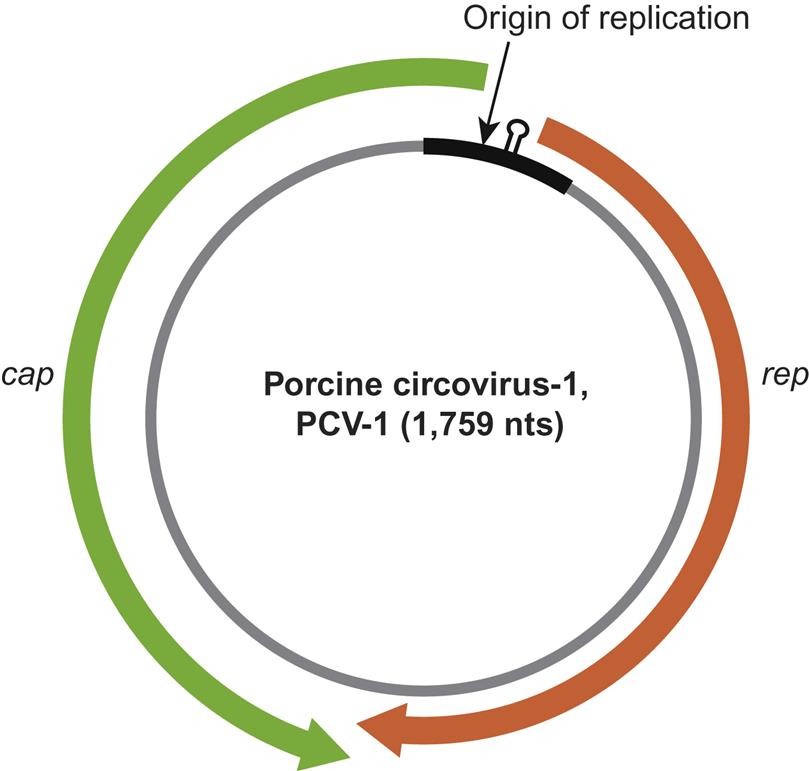
The virions of circoviruses are small (approximately 20–25 nm in diameter), nonenveloped, spherical in outline, with T=1 icosahedral symmetry (Table 13.1). Virions are made up of 60 capsid subunits that package the viral circular single-stranded DNA. Virions of individual circoviruses differ in their surface structure and morphology, with chicken anemia virus having 12 trumpet-like structures that are less obvious in the other circoviruses (Fig. 13.1). Specifically, virions of the members of the genus Circovirus have a comparatively smooth and featureless surface as compared to those of the genus Gyrovirus (ie, chicken anemia virus). Mature virions occur either as free particles within infected cells contained in diagnostic specimens, or in a linear “strings of pearls” pattern in cell-free specimens. The genome of circoviruses consists of a single molecule of circular (covalently closed ends) single-stranded ambisense (genus Circovirus) or negative sense (genus Gyrovirus) DNA, approximately 1.7–2.3 kb in size. Circoviruses are the smallest DNA viruses known to infect mammals, and their small genome is reduced to the absolute minimum to fulfill just two basic functions, ie, copying and packaging of the viral genome.
Table 13.1
Virions are small (20–25 nm), nonenveloped, spherical in outline, with icosahedral symmetry
Mature virions are present in infected cells, and they occur in linear arrays in cell-free diagnostic specimens
The genome consists of a single molecule of circular (covalently closed ends) single-stranded ambisense (genus Circovirus) or positive-sense (genus Gyrovirus) DNA, 1.7–2.3 kb in size
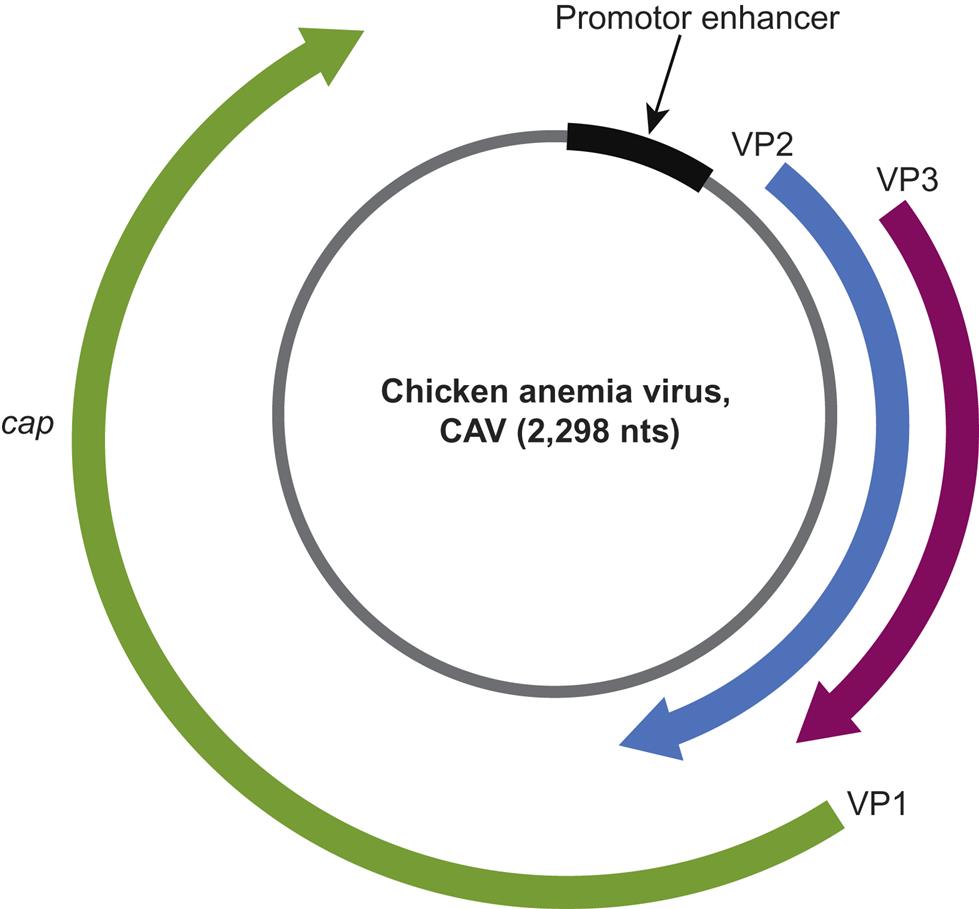
Chicken anemia virus encodes a protein (VP3) that induces apoptosis in chicken lymphocytes (apoptin)
Replication takes place in the nucleus of cycling cells. Large cytoplasmic inclusion bodies are characteristic
Virions are very stable, resisting 60°C for 30 minutes and pH 3 to pH 9
Beak and feather disease virus, porcine circoviruses 1 and 2, canine circovirus, and the other members of the genus Circovirus all utilize an ambisense transcription strategy—ie, some genes are encoded in the viral sense DNA and others in the complementary strand (Fig. 13.2). The genome of viruses in the genus Circovirus contains two major open reading frames (ORFs) oriented in the opposite direction (ambisense), one encoding the replicase protein (Rep) and the other encoding the immunogenic capsid protein (Cap). Alternative splicing is used to transcribe Rep and Rep’ RNA transcripts. The origin of virus replication (Ori) is the intergenic region between the 5′ ends of the rep and cap genes. In contrast, the genes of chicken anemia virus (genus Gyrovirus) are all encoded in the complementary positive-sense DNA strand that is transcribed to give a single polycistronic transcript (Fig. 13.3). Chicken anemia virus has three partially overlapping ORFs. The ORF1 encodes the VP1 capsid protein that is present in virions, and ORF2 and ORF3 encode the VP2 and VP3 nonstructural proteins, respectively. The VP3 protein, termed apoptin, induces apoptosis of T lymphocytes and is probably important to the pathogenesis of infections in chickens.
Circoviruses are all highly stable in the environment and, like parvoviruses, are notoriously difficult to inactivate, For example, they are not inactivated by heating at 60°C for 30 minutes, are highly resistant to many disinfectants, and may require long exposure even to efficacious chemical sterilizers.
Virus Replication
The receptors responsible for cellular attachment of circoviruses are not well characterized, but some circoviruses hemagglutinate erythrocytes and thus they are likely to bind to sialic acid residues on the cell surface. Porcine circovirus 2 utilizes heparin/heparan sulfate and chondroitin sulfate B glycosaminoglycans as general attachment receptors, which may explain why the virus exhibits broad tropism to multiple organs and tissues in pigs. Despite the ubiquitous nature of these receptors, however, cells of the monocyte/macrophage lineage are preferentially targeted. A specific entry receptor for porcine circovirus 2 has not been identified. Virus particles are taken up into cells by endocytosis, likely via clathrin-mediated endocytosis although the specific mechanisms also are not well understood. Porcine circovirus 2 is internalized by both mature and immature dendritic cells, including blood dendritic cells, plasmacytoid dendritic cells, and dendritic cell precursors, suggesting that a nonmacropinocytic uptake of the virus may be involved in virus entry. Epithelial cells are also major targets for porcine circovirus 2 in swine, and a dynamin- and cholesterol-independent but actin- and small GTPase-dependent pathway facilitates the entry and internalization of the virus into epithelial cells prior to replication.
After entry and localization of porcine circovirus 2 in endosomes, a serine protease is required for virus release from the endosome, which suggests that proteolytic cleavage of the capsid protein (Cap) occurs during the uncoating process. Viral DNA replication occurs in the nucleus of infected cells and requires cellular proteins and other components produced during the S phase of the cell cycle. Replication of the genome is believed to occur via a rolling circle that originates at a stem-loop structure. Three conserved rolling-circle replication motifs (RCR-I, RCR-II, and RCR-III) and a dNTP-binding motif are present within the replicase (Rep and Rep’) proteins of porcine circoviruses, and mutation of conserved motifs negatively affects virus replication. Three distinct proteins (VP1, VP2, and VP3) are produced during replication of chicken anemia virus. A 3-amino acid motif in VP1 is associated with rolling-circle replication. The phosphatase activity of VP2 is important, though not required, for virus replication, and VP3 is essential for completion of the virus’ life cycle.
A major feature of the circoviruses that determines their pathogenesis is the requirement for actively dividing cells to facilitate replication of their DNA, thus virus replication occurs in actively dividing cells in the tissues of young animals. Similarly, replication of porcine circovirus 2 in swine is enhanced during periods of immune stimulation that result in proliferation of lymphocytes in which the virus can replicate. Circoviruses typically cause persistent infections of their respective hosts, although the mechanisms responsible for establishing persistent infection are poorly understood as these viruses persist despite apparently robust host antiviral immune responses. In the case of chicken anemia virus, virus replication in the oviduct of chickens may be regulated by estrogen, and hence is differentially stimulated, particularly during egg laying, to allow more efficient vertical transmission. The apoptin protein of chicken anemia virus may itself cause destruction of infected lymphocytes, and hence promote a relative immune suppression that favors virus persistence.
Members of the Genus Circovirus
Beak and Feather DISEASE VIRUS
It had long been known that many species of Australian parrots undergo permanent loss of feathers and develop beak and claw deformities when in captivity. Thin-section electron microscopic examination of affected tissues from such birds undertaken in 1984 revealed large numbers of virions that resembled the previously described porcine circovirus 1. The beak and feather disease virus, or similar circoviruses, infects more than 60 different species of New World, Old World, and South Pacific psittacine birds. Circoviruses or their DNA have also been detected in many other birds, including canaries, ostriches, pigeons, ducks, geese, finches, gulls, ravens, pheasants, jays, and starlings.
Clinical Features and Epidemiology
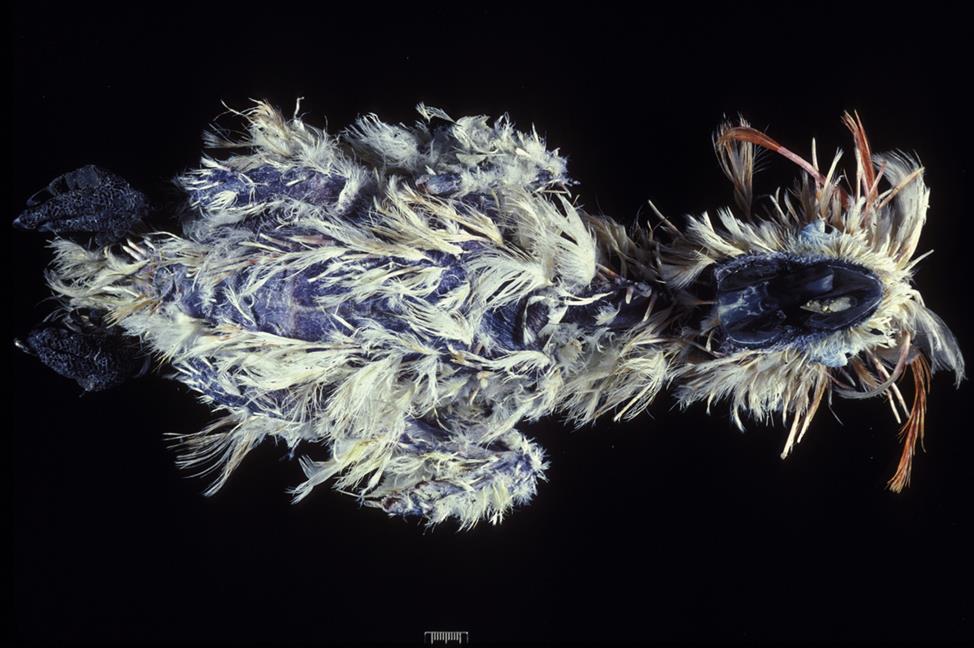
Many circovirus infections in psittacine birds are mild or subclinical. Where it occurs, beak and feather disease is a debilitating disease of cockatoos, parrots, and budgerigars, but it is principally a disease of cockatoos. Natural infection occurs primarily in birds less than 5 years of age, most often in young birds during first feather formation, although older birds can also be infected. Generally there is a high morbidity but low mortality associated with circovirus infection in birds. The prevalence of beak and feather disease virus infection is high (up to 95%) among free-ranging psittacine birds in parts of Australia but relatively low (5%) among captive birds in the United States. The natural routes of exposure are via aerosolized virus particles or direct ingestion of contaminated materials. The virus spreads through virus shedding in feather dander and also by fecal shedding and the feeding of chicks with regurgitated crop contents. Mortality and clinical signs are variable and dependent on the age, species, and concurrent infection status of susceptible birds. Typical clinical findings include feather loss, abnormal pin feathers (constricted, clubbed, or stunted), abnormal mature feathers (retention of sheaths, blood in shaft, fracture of rachis), and various beak abnormalities (Fig. 13.4). The beaks of affected birds are variously described as being shiny, overgrown, or broken, exhibiting delaminations, or with palatine necrosis. Birds may have feather lesions, beak lesions, or both. Severe leukopenia and nonregenerative anemia have been reported in some parrots, but usually without feather lesions.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree