Arteriviridae and Roniviridae
Abstract
This chapter describes the properties of arteriviruses and roniviruses, and features of the diseases they cause in animals.
Keywords
Arteriviridae; Roniviridae; arterivirus; ronivirus; Okavirus
Viruses within the families Arteriviridae and Roniviridae are included in the order Nidovirales, along with those viruses in the families Coronaviridae and Mesoniviridae (see Chapter 24: Coronaviridae). The Arteriviridae and Coronaviridae include a large group of viruses that infect vertebrates (principally mammalian viruses), whereas the Roniviridae and Mesoniviridae include viruses that infect invertebrates—crustaceans and insects, respectively. Viruses in these families have very different virion morphology, but the grouping reflects their common and distinctive replication strategy that utilizes a nested set of 3′ coterminal subgenomic messenger RNAs (mRNAs). The name of the family Arteriviridae is derived from the disease caused by its prototype species, equine arteritis virus. The family Roniviridae contains several genotypes of gill-associated and yellow head viruses.
Properties of ARTERIVIRUSES and RONIVIRUSES
Classification
The family Arteriviridae currently comprises a single genus, Arterivirus, which contains all member viruses: equine arteritis virus, porcine reproductive and respiratory syndrome virus, lactate dehydrogenase-elevating virus, simian hemorrhagic fever virus, and provisionally, wobbly possum disease virus, a novel nidovirus that was identified recently in Australian brushtail possums (Trichosurus vulpecula) in New Zealand (Table 25.1). It has been proposed that the family Arteriviridae be further subdivided taxonomically to accommodate the recently identified, highly divergent arteriviruses of African nonhuman primates and rodents. Five genera are included in this proposed classification, based on sequence and phylogenetic analysis of the open reading frame 1b. The family Roniviridae currently contains a group of related viruses causing disease in crustaceans that are members of a single genus, Okavirus.
Table 25.1
| Virus | Host | Disease |
| Equine arteritis virus | Horse | Systemic influenza-like disease, arteritis, abortion, pneumonia in foals |
| Porcine reproductive and respiratory syndrome virus | Swine | Porcine reproductive and respiratory syndrome, systemic disease; abortion of sows or birth of stillborn or mummified fetuses; respiratory disease |
| Lactate dehydrogenase-elevating virus | Mice | Usually none, but the presence of the virus may confound research using infected mice |
| Simian hemorrhagic fever virus | Macaques | Systemic hemorrhagic disease, death |
| Wobbly possum disease virus | Australian brushtail possum | Neurologic disease |
Virion Properties
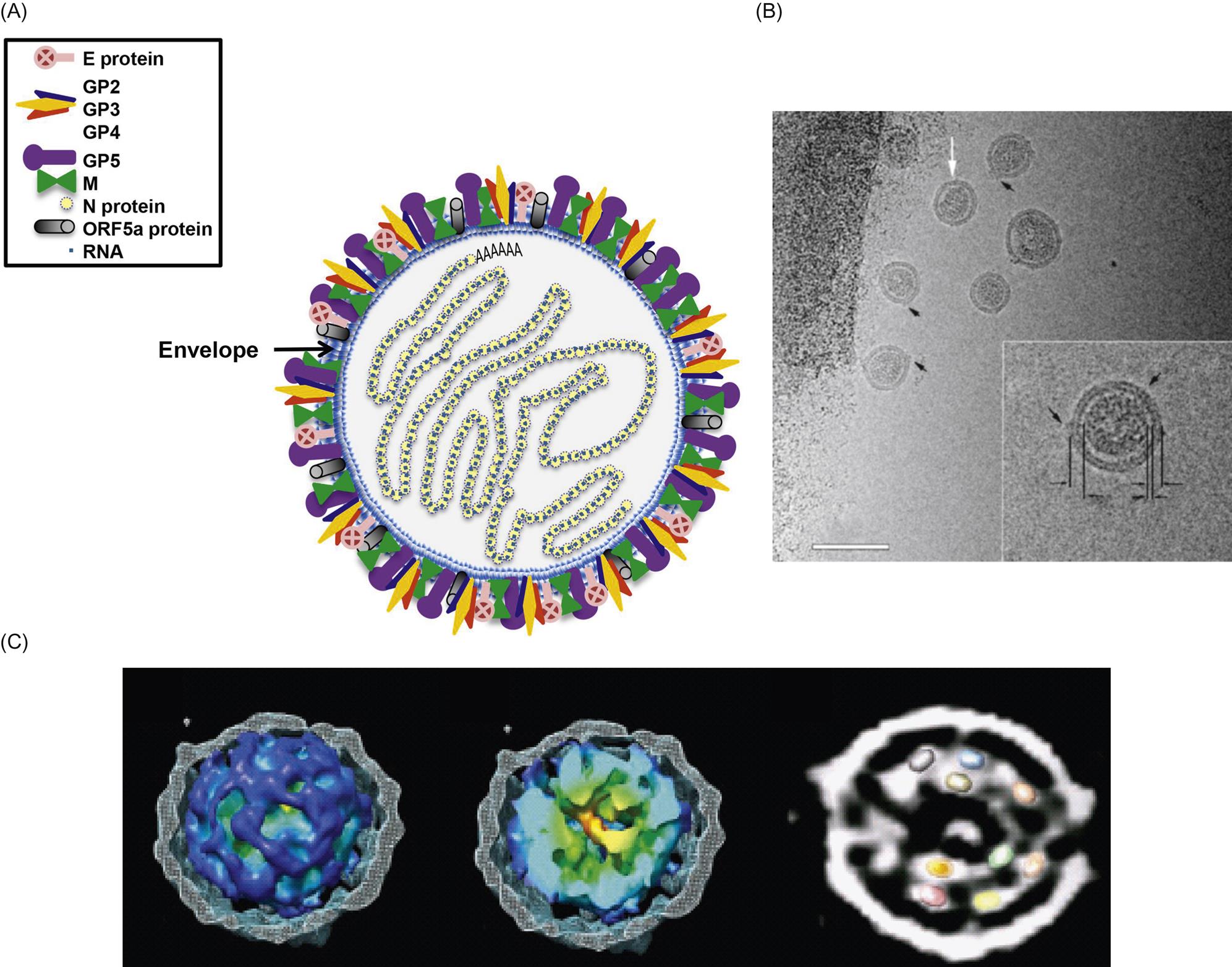
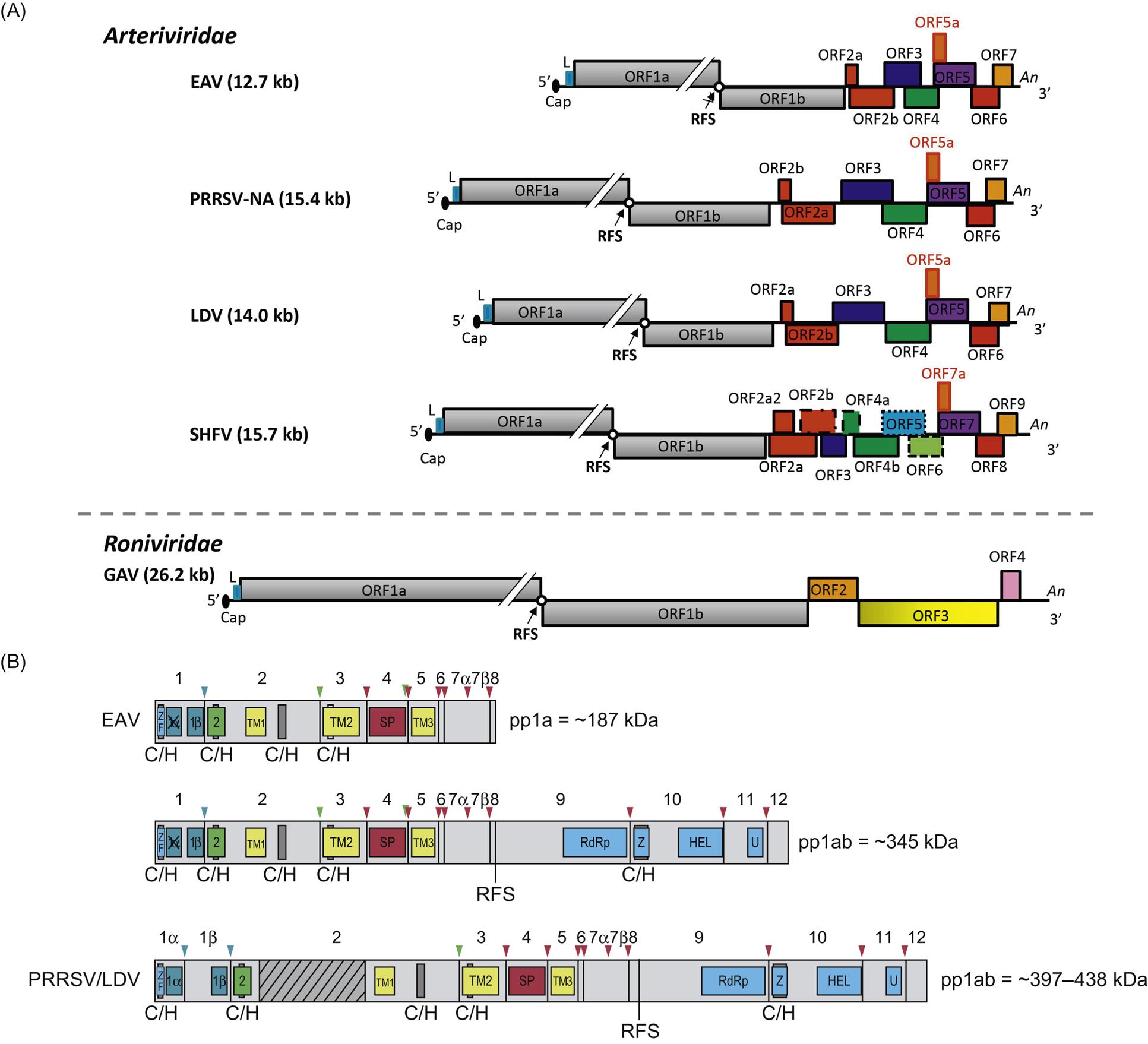
Arterivirus virions are enveloped, spherical, and 45–60 nm in diameter, which is only about half the size of those of coronaviruses (Fig. 25.1; also see Fig. 24.1). In contrast to the nucleocapsids of coronaviruses and roniviruses, which are helical, arterivirus nucleocapsids are isometric, 25–35 nm in diameter. Whereas envelope glycoprotein spikes are prominent on coronaviruses and roniviruses, they are small and indistinct on arterivirus virions. The genome of arteriviruses consists of a single molecule of linear positive-sense, single-stranded RNA, approximately 12.7–15.7 kb in size that includes 9–12 open reading frames (Fig. 25.2). There are untranslated regions at the 5′ and 3′ ends of the genome (156–224 and 59–177 nt, respectively), and a 3′-poly(A) terminal sequence. Arterivirus virions include a single nucleocapsid protein (N) and seven envelope proteins, designated E, GP2, GP3, GP4, ORF5a protein, GP5, and M. Of these, three minor envelope proteins (GP2, GP3, and GP4) form a heterotrimer, and the nonglycosylated triple-membrane spanning integral membrane protein, M, and the large envelope glycoprotein, GP5, form a heterodimer. The major neutralization determinants of arteriviruses are expressed on GP5, although the M protein exerts a conformational influence on GP5. The structural proteins of simian hemorrhagic fever virus are not as well characterized as that of the other arteriviruses, and its genome includes four additional open reading frames that may represent reduplications of genes encoding minor structural viral proteins (the reduplicated proteins being designated as E′, GP2′, GP3′, and GP4′).
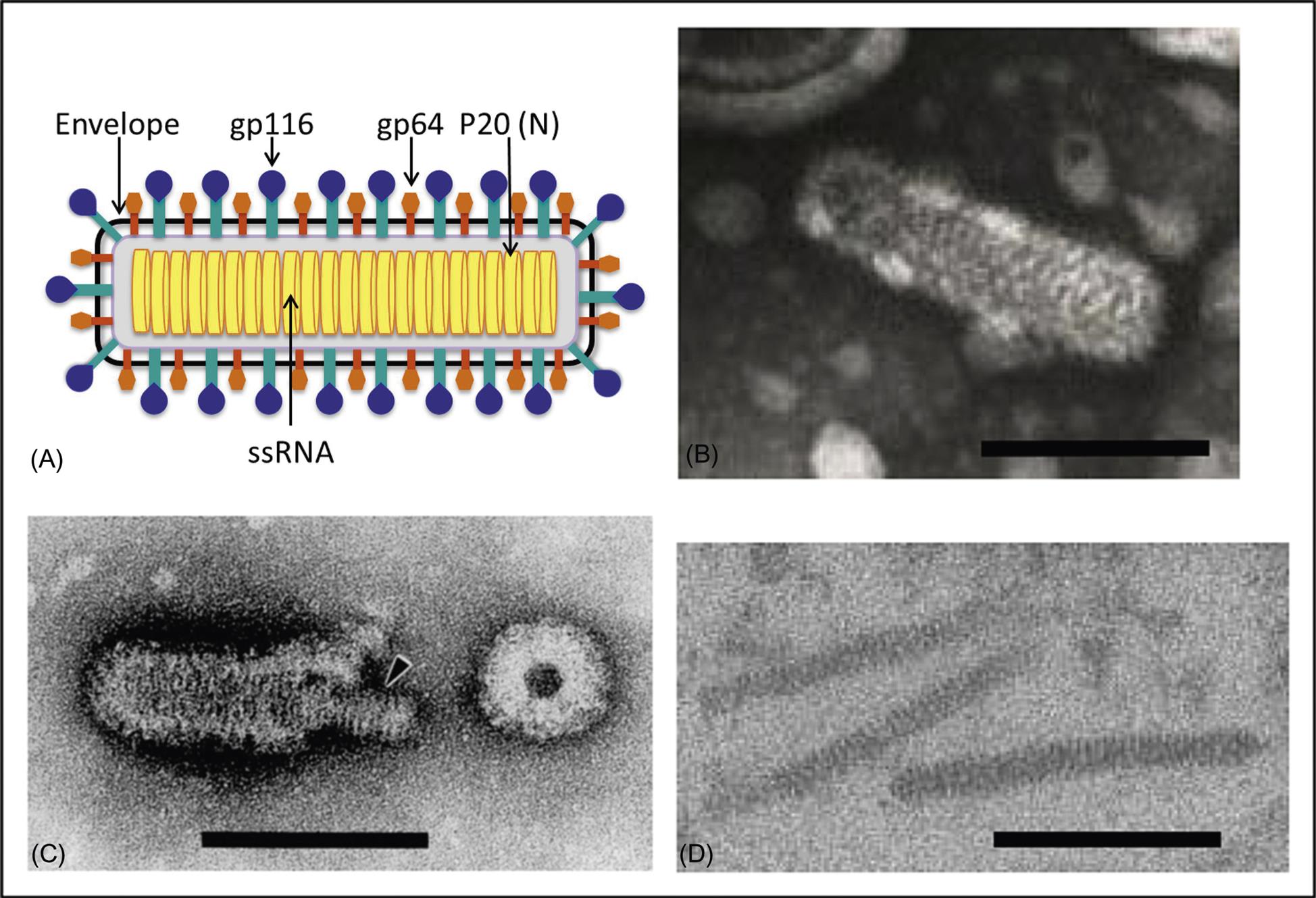
Ronivirus virions are bacilliform, 40–60 nm×150–200 nm, with rounded ends and prominent glycoprotein envelope spikes (Fig. 25.3; also see Fig. 24.1). The nucleocapsid has helical symmetry and is comprised of a coiled filament 16–30 nm in diameter. The nucleocapsid is surrounded by the envelope, which has diffuse projections (approximately 8 nm thick and 11 nm in length) extending from the surface. Like other nidoviruses, the ronivirus genome is a positive-sense, single strand RNA molecule of 26.2–26.6 kb that contains a 5′ cap structure, a 3′-poly(A) tail, and includes five open reading frames (Fig. 25.2; also see Fig. 24.2). Virions consist of at least three envelope proteins (gp116, gp64, and an N-terminal triple-membrane spanning protein of unknown function). The envelope proteins are cleavage products of a larger polyprotein precursor.
Virus Replication
The host range of arteriviruses is highly restricted, and all arteriviruses share the capacity to establish asymptomatic prolonged or persistent infections in their respective natural hosts. Arteriviruses replicate in macrophages and a very limited number of other cell types within their respective hosts. Equine arteritis virus is less fastidious than other arteriviruses as it replicates in vitro in a variety of primary cell cultures, including equine pulmonary artery endothelial, horse kidney, rabbit kidney, and hamster kidney cells, and in a wide variety of cell lines such as baby hamster kidney (BHK-21) and rabbit kidney (RK-13). Other arteriviruses (eg, porcine reproductive and respiratory syndrome, lactate dehydrogenase elevating, and simian hemorrhagic fever viruses) typically replicate in only cultured macrophages or a very limited number of other cell types and/or lines. Some arteriviruses effectively can subvert protective host innate immune responses, including apoptosis of infected macrophages and interferon signaling pathways.
Similar to other enveloped viruses, arteriviruses bind to cell surface receptor(s) using their envelope proteins to mediate the process of cell attachment and fusion with the host cell membrane. The receptors for most arteriviruses are uncharacterized; however, potential receptors involved in the attachment and internalization of porcine reproductive and respiratory syndrome virus include CD163 (a cellular protein in the scavenger receptor cysteine-rich superfamily), sialoadhesin (or sialic acid-binding immunoglobulin (Ig)-like lectin 1 (CD169)), and heparan sulfate glycosaminoglycans. Recent studies using recombinant chimeric viruses confirm that the ectodomains of the major envelop proteins GP5 and M are not the essential determinants of cellular tropism of equine arteritis virus, rather the heterotrimer of minor envelope proteins GP2, GP3, and GP4 is responsible for cell tropism and receptor binding. Arteriviruses appear to enter susceptible cells by a low-pH-dependent endocytic pathway.
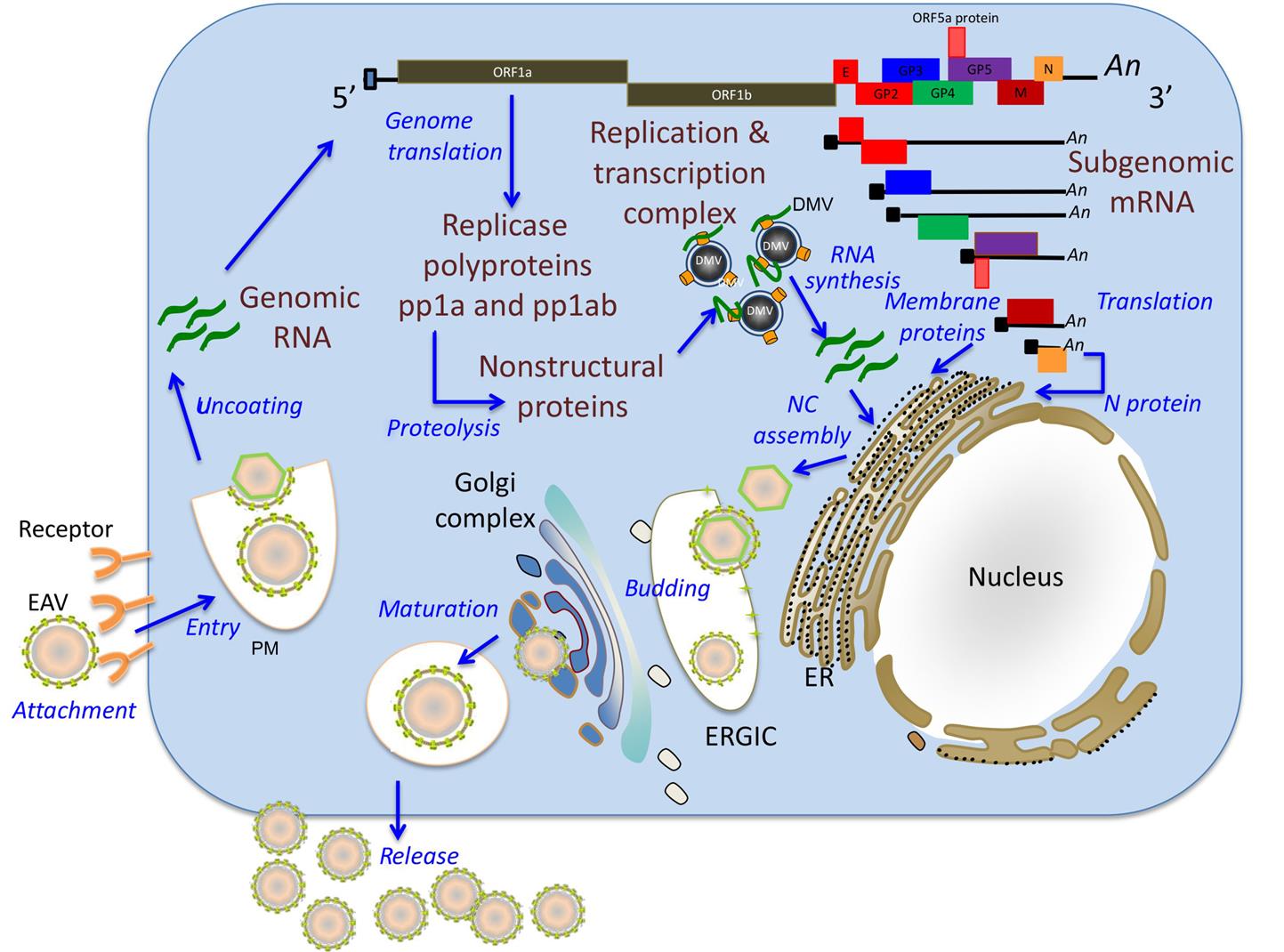
Similar to other nidoviruses, replication and transcription of arteriviruses are processed through different minus-strand intermediates: a full-length minus-strand RNA template (or antigenome) is used for replication, while subgenome-sized minus strands produced during a process of discontinuous RNA synthesis are used to synthesize subgenomic mRNAs (Fig. 25.4). The two large open reading frames at the 5′ end of the arterivirus genome encode two replicase polyproteins that are expressed directly from viral genomic RNA through a ribosomal frameshifting mechanism. These replicase polyproteins are co- and post-translationally modified by viral proteinases into at least 13 (equine arteritis virus) or 16 (porcine reproductive and respiratory syndrome virus) nonstructural proteins (nsps) that mediate replication. The genes that encode the viral structural proteins are overlapping, and located in the 3′ end of the genome; they are expressed from a nested set of 3′ coterminal subgenomic RNAs (Fig. 25.2). These subgenomic RNAs all include a common 5′ leader sequence derived from the 5′ untranslated region of viral genomic RNA, at least one unique open reading frame encoding one or more structural virion proteins, and a common 3′-poly(A) tail. The individual open reading frames that are included in these subgenomic mRNAs reflect overlapping reading frames contained in the 3′ end of the viral genome. It is believed that the subgenomic mRNAs are generated by discontinuous transcription that links noncontiguous portions of the viral genome, to produce negative-strand templates that are transcribed into positive-strand subgenomic mRNAs that are then translated into the individual virion proteins.
In arterivirus-infected cells, newly synthesized nonstructural proteins are incorporated into cellular organelles, particularly the endoplasmic reticulum, which results in membrane pairing and formation of double-membrane vesicles. Immunoelectron microscopic studies have confirmed that viral nonstructural proteins that are part of the replication/transcription complexes and nascent viral RNA are associated with these double-membrane vesicles suggesting that these vesicles carry the enzyme complex responsible for virus replication and subgenomic mRNA synthesis. Arterivirus replication occurs in the cytoplasm of infected cells, although nonstructural (nsp1) and structural (N) proteins selectively translocate to the nucleus. During the early stages of infection, nsp1 is primarily located in the nucleus while its perinuclear cytoplasmic localization becomes evident later in infection. The newly synthesized genome is encapsidated into the N protein to form the nucleocapsid, which becomes enveloped by budding through the endoplasmic reticulum–Golgi intermediate compartment that contains membranes that include the viral envelope proteins. Newly formed virions mature in the Golgi complex during their movement through the exocytic pathway and are ultimately released from infected cells (Table 25.2).
Table 25.2
Virions are spherical, 50–70 nm in diameter, with an isometric nucleocapsid and a closely adherent smooth-surfaced envelope with ring-like structures
The genome consists of a single molecule of linear, positive-sense, single-stranded RNA, 13–15 kb in size. Virion RNA has a 5′ cap and its 3′ end is polyadenylated; the genomic RNA is infectious
Replication takes place in the cytoplasm; the genome is transcribed to form a full-length negative-sense RNA, from which is transcribed a 3′ coterminal nested set of mRNAs; only the unique sequences at the 5′ end of each mRNA are translated
Virions are formed by budding into the endoplasmic reticulum, from where they are released by exocytosis
The overall strategy for replication of roniviruses is similar to that of arteriviruses, but the structural proteins are expressed from a nested set of just two subgenomic mRNAs that each encode several proteins. The subgenomic mRNAs also lack a common 5′ leader sequence derived from genomic viral RNA.
Members of the Family Arteriviridae, Genus Arterivirus
EQUINE ARTERITIS Virus
Descriptions of a disease that very probably was equine viral arteritis were first published in the late 18th and early 19th centuries, with colloquial names such as “pinkeye,” “infectious or epizootic cellulites,” “influenza erysipelatosa,” and “Pferdestaupe.” Early investigators also recognized that apparently healthy stallions transmitted this disease to susceptible mares at breeding. The causative agent, equine arteritis virus, was first isolated in 1953 from lung tissue of an aborted fetus during an epizootic of abortion and respiratory disease on a breeding farm near Bucyrus, Ohio. After isolation of the causative virus and description of characteristic vascular lesions, equine viral arteritis was identified as an etiologically distinct disease of the horse. Equids are the only known natural host of equine arteritis virus.
Although serologic studies indicate that infection occurs worldwide, the incidence of both infection and overt disease varies markedly between countries and among horses of different breeds. Serologic surveys confirm equine arteritis infection of horses in North and South America, Europe, Australia, Africa, and Asia, whereas other countries such as Iceland and Japan are apparently free of the virus. New Zealand recently eradicated equine arteritis virus infection.
Clinical Features and Epidemiology
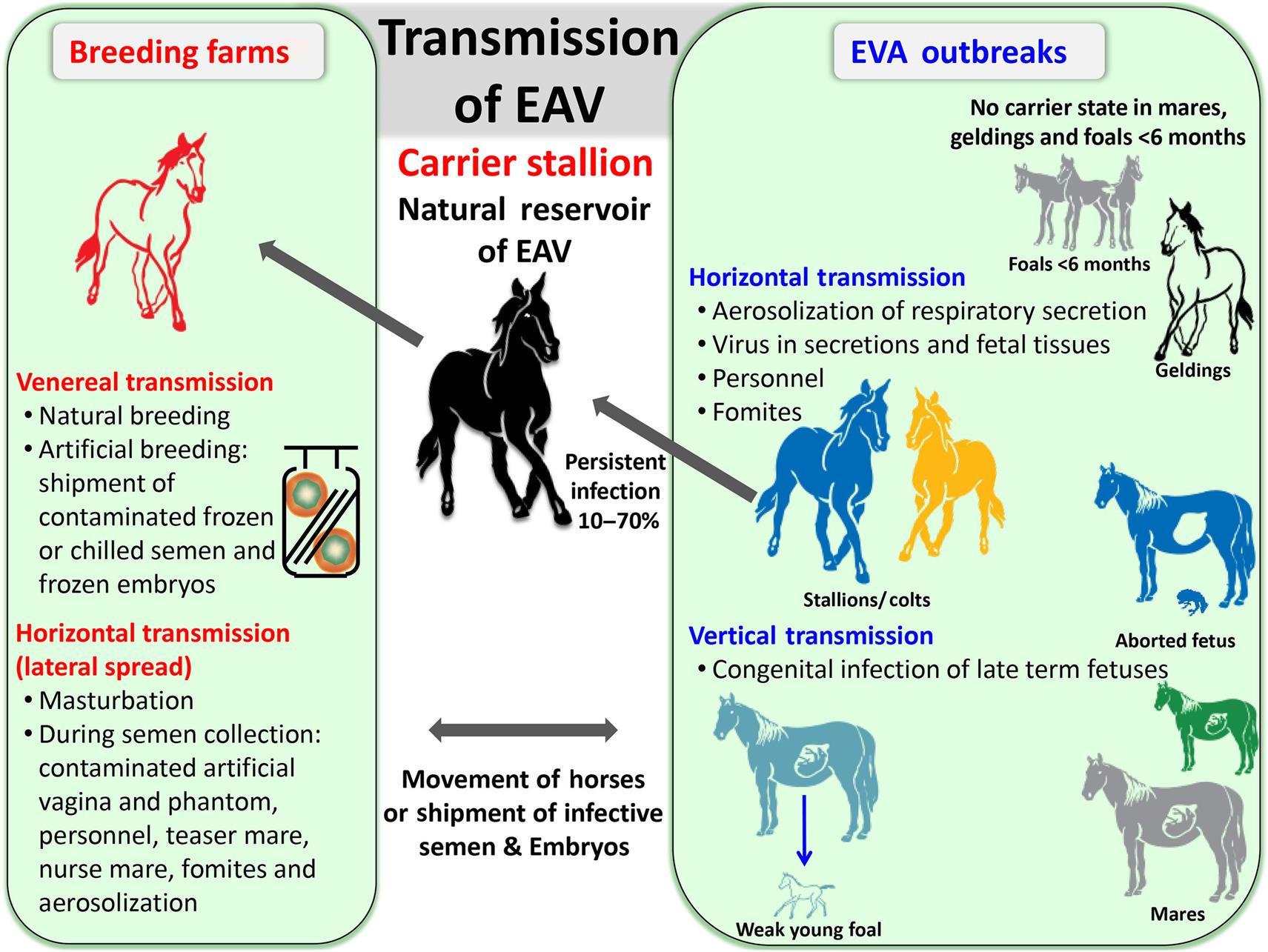
Most natural infections with equine arteritis virus are subclinical or mild, and descriptions of fatal disease are based on experimental infections with highly horse-adapted laboratory strains of the virus. Nevertheless, relatively virulent field strains of equine arteritis virus periodically cause natural outbreaks of equine viral arteritis in horses. After an incubation period of 3–14 days (typically 6–8 days following venereal exposure), the onset of disease is marked by fever (>41°C), leukopenia, depression, excessive lacrimation, anorexia, conjunctivitis, rhinitis and nasal discharge, urticaria of the head, neck, and trunk, and edema, which is most pronounced over the eyes (supraorbital), the abdomen, including the prepuce, scrotum, and mammary glands, and the hind limbs (often resulting in a stiff gait). Although naturally infected horses usually recover uneventfully, death as a result of rapidly progressive bronchointerstitial pneumonia occurs sporadically in young (neonatal) foals, and a progressive “pneumo-enteric” syndrome is described in 1–3 month-old foals. Abortion is characteristic of infections of pregnant mares with particular strains of the virus, and infection of large numbers of susceptible (unvaccinated) pregnant mares can lead to “abortion storms.” Abortion generally occurs 10–30 days after infection and at any time between 3 and 10 months of gestation; it is linked closely with the late febrile or early convalescent phase of infection, but can occur even if no clinical signs are noticed. A variable proportion of acutely infected stallions (10–70%) become persistently infected and shed the virus exclusively in their semen. Persistent infection is maintained in the reproductive tract of individual stallions for variable intervals, from several weeks to lifelong. There is no compelling evidence of equine arteritis virus causing persistent infection in mares, geldings, or foals.
Equine arteritis virus is spread by both the respiratory and venereal routes, respectively by aerosol from acutely infected horses or in the semen of persistently infected (carrier) stallions (Fig. 25.5). The latter are the essential natural virus reservoir, and play an important role in maintenance and perpetuation of equine arteritis virus in nature. Virus is spread from carrier stallions exclusively via the venereal route; semen collected from persistently infected stallions and used in artificial insemination has been responsible for outbreaks of disease. Furthermore, genetic diversity is generated in equine arteritis virus during persistent infection of stallions.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree