Arenaviridae*
Abstract
This chapter describes the properties of arenaviruses and features of the diseases they cause in animals.
Keywords
Arenaviridae; arenavirus; virus host ecology; disease pathogenesis
The arenaviruses are a genetically diverse and broadly distributed group of viruses, some of which are significant pathogens of humans and, more recently, reptiles (snakes). Ancient coevolutionary relationships with rodent reservoir hosts are a fundamental determinant of the ecology of zoonotic arenavirus infections. The risk of transmission of each of the zoonotic arenaviruses to humans relates to the nature of the infection of its rodent host (usually persistent asymptomatic infection with lifelong virus shedding), rodent population dynamics and behavior, in addition to human occupational and other risk factors that lead to exposure to virus-laden rodent excreta. Human infection can lead to some of the most lethal hemorrhagic fevers known: Lassa, Lujo, Argentine (Junin virus), Bolivian (Machupo virus), Venezuelan (Guanarito virus), Brazilian (Sabiá virus), and Chapare hemorrhagic fevers. The viruses that cause these human diseases are BioSafety Level 4 pathogens; they must be handled under maximum containment conditions to prevent human exposure in the laboratory. The recently identified boid inclusion body disease complex of viruses (University of Helsinki virus, Golden Gate virus, and others) isolated from diseased boa constrictor snakes has expanded the host range of arenaviruses to include not only mammals but also reptiles. These reptile-associated viruses are not known to cause human disease, and their discovery highlights important gaps in the understanding of arenavirus ecology and pathogenesis in nonmammalian hosts.
The prototype arenavirus, lymphocytic choriomeningitis virus, has over many years played two disparate roles in comparative virology: wild-type strains of the virus are zoonotic pathogens and the subject of public health surveillance in some countries where they are enzootic, whereas laboratory strains have provided much of the conceptual basis for current understanding of viral immunology and pathogenesis using laboratory mouse models.
Properties of ARENAVIRUSES
Classification
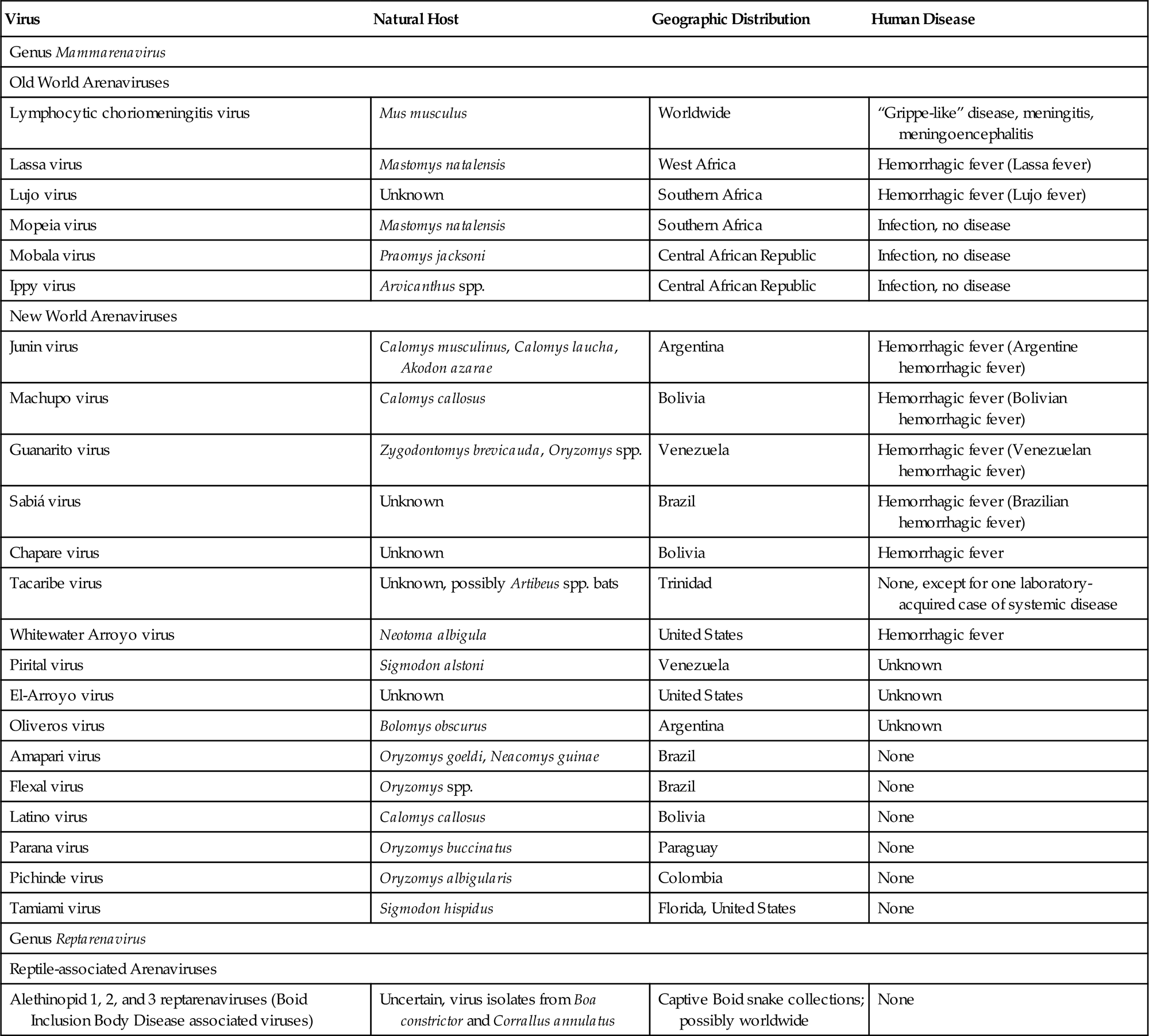
The family Arenaviridae contains two genera, namely Mammarenavirus with 27 virus species, and Reptarenavirus with three virus species (Table 23.1). Within the genus Mammareniavirus are the Old World arenaviruses, including lymphocytic choriomeningitis virus (now designated lymphocytic choriomeningitis mammarenavirus), which is allied with the common house mouse, Mus musculus. This virus is now enzootic throughout the world because of global colonization by Mus musculus. Other Old World arenaviruses are associated with Mastomys spp. and Praomys spp. rodents or other as yet unidentified reservoirs in Africa. The African viruses include Lassa virus and Lujo virus (respectively, Lassa mammarenavirus and Lujo mammarenavirus), both of which produce severe disease in humans, and a few other viruses that infect humans but are not known to cause disease. A second subgroup within the genus Mammarenavirus includes the New World arenaviruses (previously also called the Tacaribe complex), which are associated with many different rodents in North, Central, and South America. This subgroup, which is further subdivided into three distinct clades, contains the important human pathogens Chapare, Guanarito, Junin, Machupo, and Sabiá viruses (now designated as Chapare Guanarito, Junin, Machupo, and Sabiá mammarenaviruses), and several other viruses that are pathogenic for humans. A group of novel arenaviruses (alethinopid 1, 2 and 3 reptarenaviruses) associated with severe disease in reptiles (inclusion body disease of boa constrictor [boid] snakes) is included in the genus Reptarenavirus.
Table 23.1
Natural History and Zoonotic Disease Potential of Arenaviruses
| Virus | Natural Host | Geographic Distribution | Human Disease |
| Genus Mammarenavirus | |||
| Old World Arenaviruses | |||
| Lymphocytic choriomeningitis virus | Mus musculus | Worldwide | “Grippe-like” disease, meningitis, meningoencephalitis |
| Lassa virus | Mastomys natalensis | West Africa | Hemorrhagic fever (Lassa fever) |
| Lujo virus | Unknown | Southern Africa | Hemorrhagic fever (Lujo fever) |
| Mopeia virus | Mastomys natalensis | Southern Africa | Infection, no disease |
| Mobala virus | Praomys jacksoni | Central African Republic | Infection, no disease |
| Ippy virus | Arvicanthus spp. | Central African Republic | Infection, no disease |
| New World Arenaviruses | |||
| Junin virus | Calomys musculinus, Calomys laucha, Akodon azarae | Argentina | Hemorrhagic fever (Argentine hemorrhagic fever) |
| Machupo virus | Calomys callosus | Bolivia | Hemorrhagic fever (Bolivian hemorrhagic fever) |
| Guanarito virus | Zygodontomys brevicauda, Oryzomys spp. | Venezuela | Hemorrhagic fever (Venezuelan hemorrhagic fever) |
| Sabiá virus | Unknown | Brazil | Hemorrhagic fever (Brazilian hemorrhagic fever) |
| Chapare virus | Unknown | Bolivia | Hemorrhagic fever |
| Tacaribe virus | Unknown, possibly Artibeus spp. bats | Trinidad | None, except for one laboratory-acquired case of systemic disease |
| Whitewater Arroyo virus | Neotoma albigula | United States | Hemorrhagic fever |
| Pirital virus | Sigmodon alstoni | Venezuela | Unknown |
| El-Arroyo virus | Unknown | United States | Unknown |
| Oliveros virus | Bolomys obscurus | Argentina | Unknown |
| Amapari virus | Oryzomys goeldi, Neacomys guinae | Brazil | None |
| Flexal virus | Oryzomys spp. | Brazil | None |
| Latino virus | Calomys callosus | Bolivia | None |
| Parana virus | Oryzomys buccinatus | Paraguay | None |
| Pichinde virus | Oryzomys albigularis | Colombia | None |
| Tamiami virus | Sigmodon hispidus | Florida, United States | None |
| Genus Reptarenavirus | |||
| Reptile-associated Arenaviruses | |||
| Alethinopid 1, 2, and 3 reptarenaviruses (Boid Inclusion Body Disease associated viruses) | Uncertain, virus isolates from Boa constrictor and Corrallus annulatus | Captive Boid snake collections; possibly worldwide | None |
Virion Properties
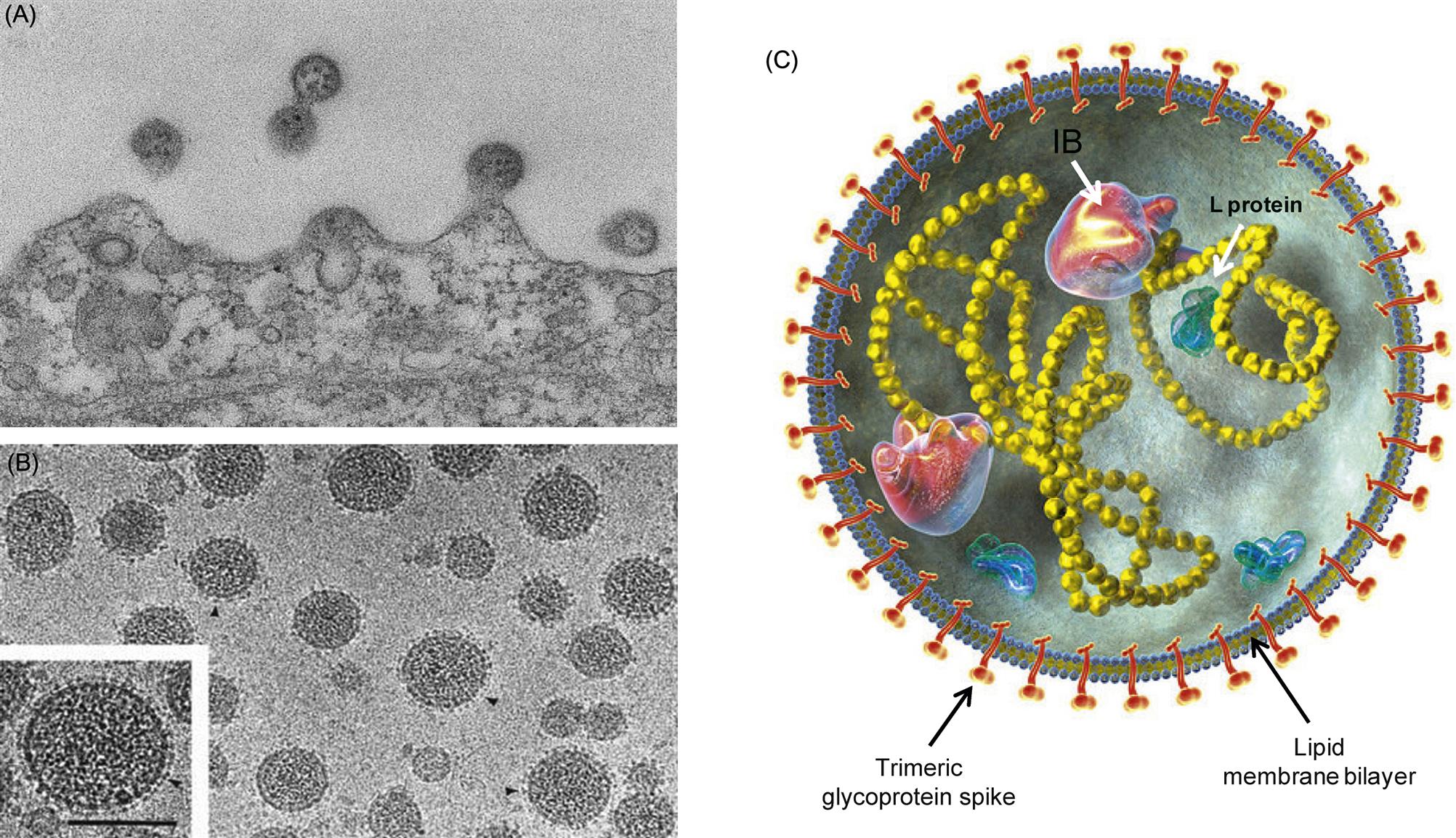
Arenavirus virions are highly pleomorphic. Their size ranges from 50 to 300 nm, although most virions have a diameter of 110–130 nm (Fig. 23.1). Virions are composed of an envelope covered with club-shaped spikes 8–10 nm in length composed of viral glycoproteins GP1 and GP2. Virions contain at least two circular helical nucleocapsid segments, each resembling a string of beads. The nucleocapsids are circular as a consequence of the genomic RNA forming “panhandles”—that is, noncovalent bonds between conserved complementary nucleotide sequences at the 3′ and 5′ ends of each RNA genome segment. The family derives its name from the presence within virions of cellular ribosomes, which, under thin section electron microscopy, resemble grains of sand (latin: arena=sand). The biological significance of this distinctive and unusual property remains uncertain. The genome of arenaviruses consists of two segments of single-stranded RNA, designated large (L) and small (S), approximately 7.5 and 3.5 kb in size, respectively. Virions may contain multiple copies of the two genome segments, often with more copies of the S RNA segment.
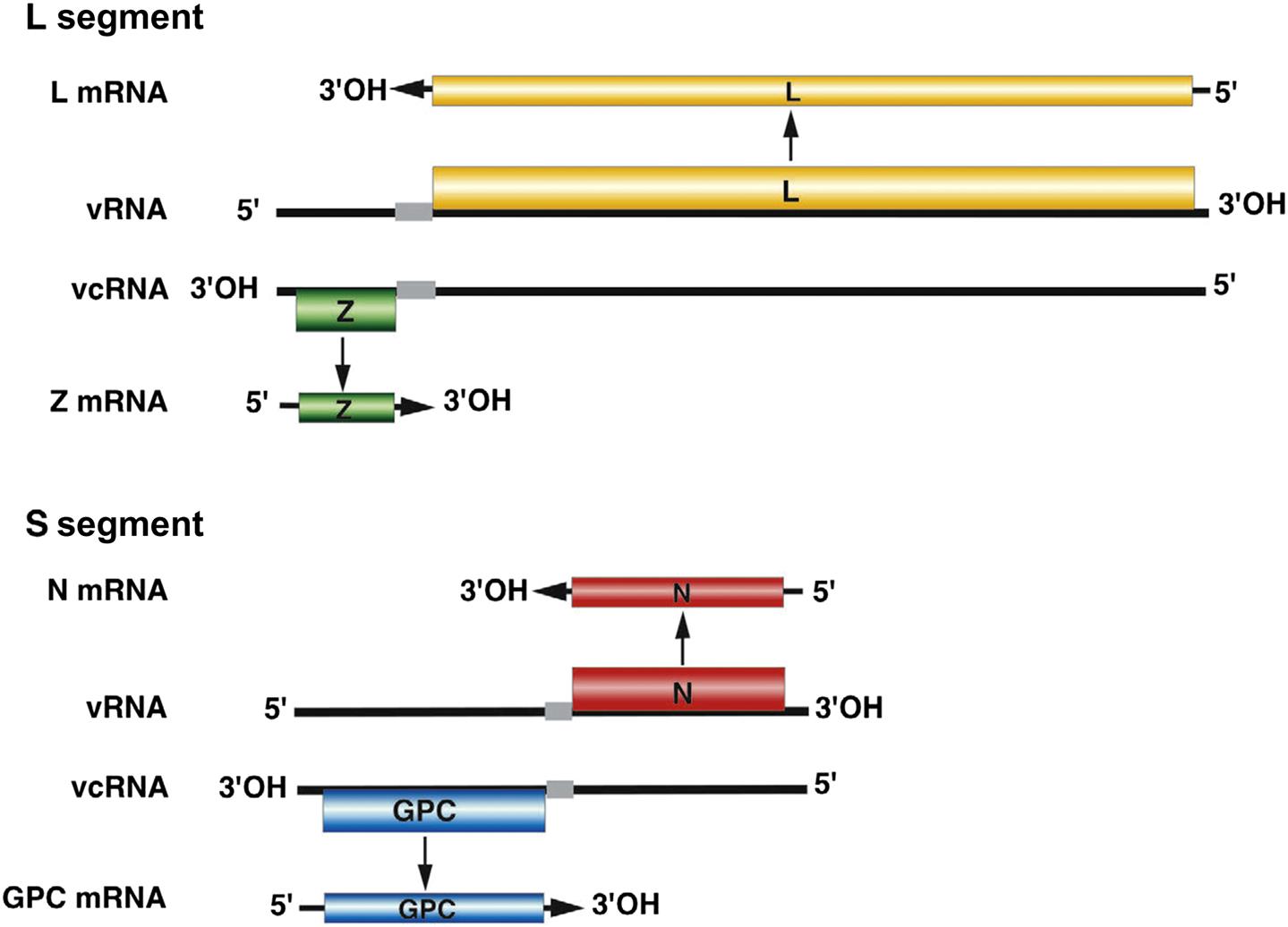
Most of the genome is of negative sense, but the 5′ half of the S segment and the 5′ end of the L segment are of positive sense; the term ambisense is used to describe this unusual genome arrangement, which is also found in some members of the family Bunyaviridae (Fig. 23.2). Specifically, the nucleoprotein (N) is encoded in the 3′ half of the complementary-sense S RNA, whereas the viral glycoprotein precursor (GPC) is encoded in the 5′ half of the viral-sense S RNA. The RNA polymerase (L) is encoded in the 3′ end of the complementary-sense L RNA and a multifunctional zinc-binding protein (Z) which also serves as a virion matrix protein is encoded in the 5′ end of the viral-sense L RNA. RNAs contain hairpin configurations between the genes (intergenic regions) that function to terminate transcription from viral and viral complementary RNAs. The mRNAs are capped—5′-methylated caps are cleaved from host mRNAs by the viral RNA-dependent RNA polymerase (transcriptase), which also has endonuclease activity, and are added to viral mRNAs to prime transcription (so-called cap snatching).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree