CHAPTER 1 The Equine Immune System
EQUINE IMMUNOLOGY
Although much of modern immunology has focused on humans and murine models of human diseases, the horse has played a significant role in our understanding of immunologic processes. These contributions include the earliest work on serotherapy and passive transfer; immunoglobulin structure and function; immunity to infectious agents; immunodeficiencies; and, more recently, reproductive immunology. Work in the horse continues in many of these areas to the benefit of equine medicine and comparative immunology. The overall organization and function of the equine immune system are similar to those of other mammalian species, although there are differences. The reader is referred to any one of a number of texts1–3 for a more in-depth review of basic immunology. Here we shall focus on those aspects of the immune system that may be of most interest to equine researchers and clinicians. When possible, pertinent references to equine work will be provided.
INNATE IMMUNITY AND THE ACUTE INFLAMMATORY RESPONSE
The horse, like every other species, is under constant assault from a variety of microbes that share its living space. Although most of these organisms are thought to be harmless, their disease-causing potential is evident when they cause opportunistic infections in individuals with compromised immune systems.4 Mammals, in general, have evolved a variety of defensive measures to prevent infections. The first line of defense includes the physical barriers provided by the skin and the mucosal surfaces of the digestive, respiratory, and urogenital tracts. In addition to providing a barrier to penetration, the surface of the skin contains various enzymes, fatty acids, and oils that inhibit the growth of bacteria, fungi, and viruses. Mucous membranes and mucosal secretions contain bacteriolytic enzymes, bacteriocidal basic polypeptides, mucopolysaccharides, and antibodies that prevent colonization and penetration of these surfaces. Mucus also provides a physical barrier that entraps invading organisms and leads to their eventual disposal.5 Particles trapped in the mucous secretions of the respiratory tract, for example, are transported upwards through the action of ciliary cells to the trachea, where they are swallowed.6 Once they are swallowed, the acidic secretions and digestive enzymes of the stomach destroy most organisms. Normal epithelial and tissue architecture is essential for successful exclusion of bacteria, and the disruption of this mechanism makes the host susceptible to infection by bacteria that normally colonize the upper airway.7,8
Acute Phase Proteins, Pro-inflammatory Cytokines, and Complement
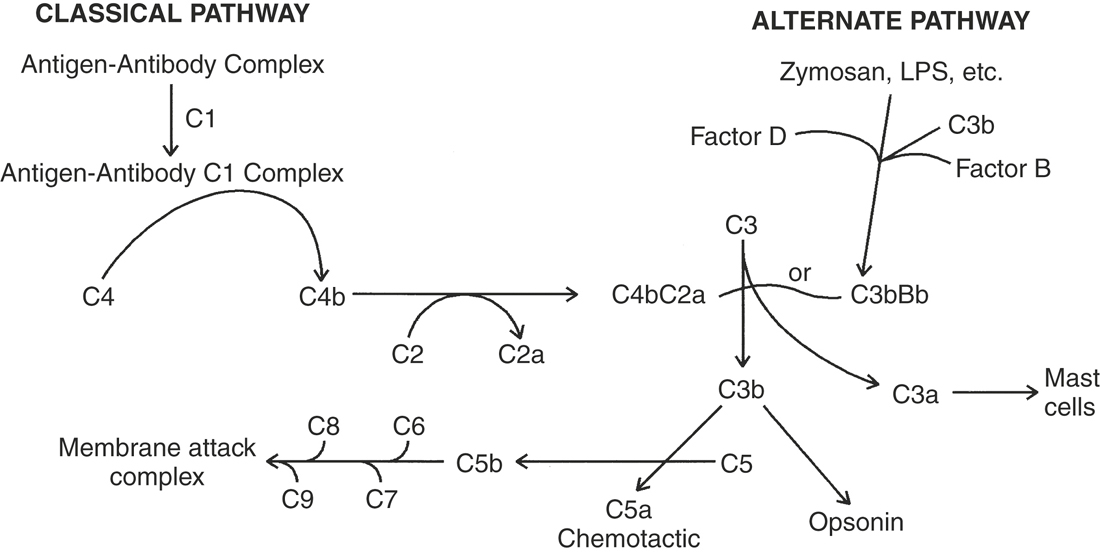
Once breached, the host presents a variety of internal defenses to contain and eliminate the invaders. Invading organisms can initiate an inflammatory response either via the activation of plasma protease systems directly, such as by bacterial cell wall components, or by the secretion of toxins or other proteins that can directly activate the inflammatory response.9 The cell walls and membranes of bacteria contain various proteins and polysaccharides with characteristic, often repeating, molecular structures. These pathogen-associated molecular patterns (PAMPs) include such molecules as lipopolysaccharides (LPSs), peptidoglycans, lipoteichoic acid, and flagellins.10 Other PAMPs include single- and double-stranded ribonucleic acid (RNA) found on viruses and unmethylated deoxyribonucleic acid (DNA) characteristic of bacteria. These PAMPs are recognized by a class of receptors known as toll-like receptors (TLRs), initially identified in Drosophila melanogaster (the fruitfly).10 This ancient family of receptors recognizing different PAMPs is widely distributed on the various cells of the body. Not surprisingly, a number of different TLRs are found on the cells of the immune system, particularly those cells involved in the initial encounter with invading microbes. The binding of a PAMP to its specific TLR leads to an intracellular signaling event, culminating in the expression of various accessory proteins that provide the co-stimulatory signals for the developing adaptive immune response. Injured cells also release products that initiate plasma protease cascades or produce pro-inflammatory cytokines that augment the inflammatory process. Resident macrophages that encounter the invader add to the genesis of the inflammatory response through the production of pro-inflammatory cytokines such as interleukin (IL)-1, IL-6, and tumor necrosis factor alpha (TNF-α).9 Cytokines are hormonelike proteins that mediate a variety of cellular responses. A vast number of cytokines are involved in the regulation of innate and adaptive immune responses. IL-1, for example, is a pleiotropic mediator of the host response to infections and injurious insults (Box 1-1). Many of the effects of IL-1 are mediated through its capacity to increase the production of other cytokines, such as granulocyte colony-stimulating factor (G-CSF), TNF-α, IL-6, IL-8, platelet-derived growth factor (PDGF), and IL-11 (cytokines, chemokines, and interleukins are discussed later). IL-6 is responsible for the increased production of acute phase proteins (Table 1-1) by hepatocytes. Although the function of some of the acute phase proteins remains unclear, many of these proteins and the cytokines that elicited them are responsible for the characteristic physical signs of inflammation, including increased blood flow and vascular permeability, migration of leukocytes from the peripheral blood into the tissues, accumulation of leukocytes at the inflammatory focus, and activation of the leukocytes to destroy any invading organisms.11 The acute phase proteins include a number of complement proteins. The complement system is an interacting series of proteases and their substrates, resulting in the production of physiologically active intermediaries that can damage membranes, attract neutrophils and other cells, increase blood flow and vascular permeability, and opsonize bacteria and other particles for phagocytosis.12 The complement cascade can be activated in two ways (Figure 1-1). The classical pathway involves the recognition and binding of C1 to antigen-antibody complexes. Bound C1 is proteolytic and cleaves C4. This cleavage of C4 leads to the binding of C2 to C4b. C2 is in turned cleaved by C1 into C2a. The C4bC2a complex is referred to as the classical pathway C3 convertase because it is a protease capable of cleaving C3 into C3a and C3b. Another C3 convertase is generated via the alternate pathway. The activation of complement via the alternate pathway does not involve antibodies; instead, certain microbial products (zymosan and LPS) stimulate the association of Factor D, a proteolytic enzyme, with the complex of Factor B and C3b leading to the formation of the C3bBb complex, which is the alternative pathway C3 convertase. C3a, produced by the cleavage of C3 by the C3 convertases can bind to mast cells, causing them to degranulate, and is thus referred to as an anaphylatoxin, as is C4a. C3b serves as an opsonin for C3b receptor-bearing phagocytic cells. C3b is also required for the formation of the membrane attack complex by the terminal complement components, C5 through C9. In this process C5 is cleaved by either the C4b2a3b (Classic pathway C5 convertase) or C3b, Bb, and properidin (alternate pathway C5 convertase). C5 is cleaved into C5a and C5b. C5a is a chemoattractive factor for neutrophils and monocytes.13 C5b forms a complex with C6, C7, and C8 on cell surfaces. This leads to the insertion and polymerization of C9 that forms a pore in the membrane, leading to cell lysis.
BOX 1-1 BIOLOGIC ACTIVITIES OF INTERLEUKIN 1
| Activates T cells | Induces fever |
| Activates B cells | Cytotoxic for some tumor cells |
| Enhances NK cell killing | Cytostatic for other tumor cells |
| Fibroblast growth factor | Stimulates collagen production |
| Stimulates PGE synthesis | Stimulates keratinocyte growth |
| Stimulates bone resorption | Stimulates mesangial cell growth |
| Chemotactic for neutrophils | Activates neutrophils |
| Activates osteoclasts | Induces IL-6 production |
TABLE 1-1 Acute Phase Proteins
| Name | Function |
|---|---|
| C3, C4, and Factor B | Opsonins |
| C-reactive protein | Opsonin and complement activator |
| Fibrinogen | Fibrin precursor, clotting factor |
| Kininogen | Kinin precursor |
| Alpha1-acid glycoprotein | Function unknown, immunomodulatory |
| Ceruloplasmin, Ferritin | Iron restriction |
| Haptoglobulin | Binds free hemoglobulin |
| Hemopexin | Binding free heme |
| Serum amyloid A (SAA) | Lipid transporter, inhibitor of neutrophil function |
| Serum amyloid P (SAP) | Lipid transporter, inhibitor of neutrophil function |
| α1-antichymotrypsin | Protease inhibitor |
| α2-macroglobulin | Protease inhibitor |
| Cysteine protease inhibitor | Protease inhibitor |
Lipid Mediators
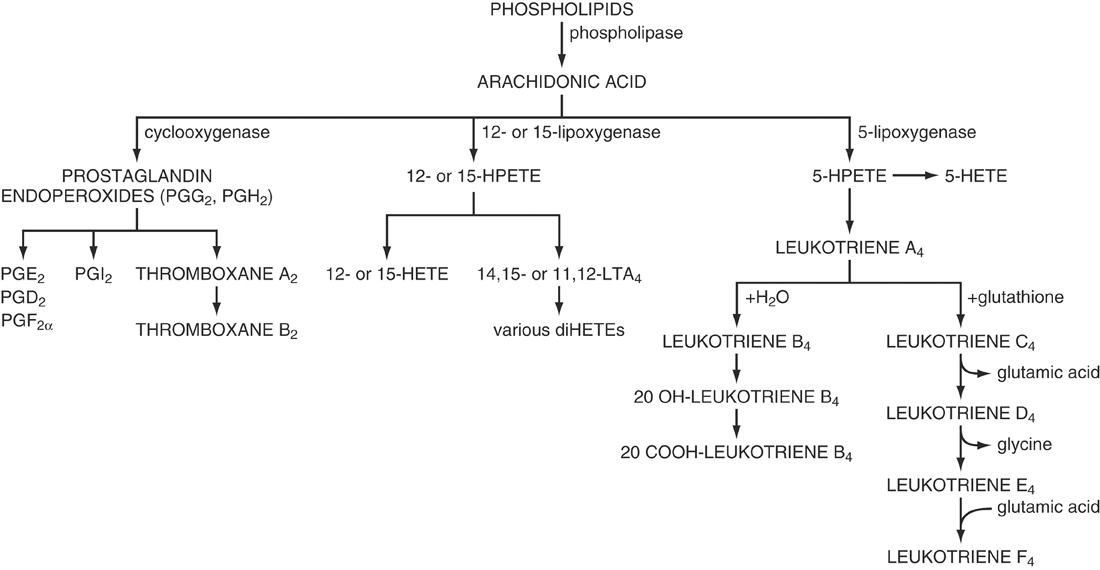
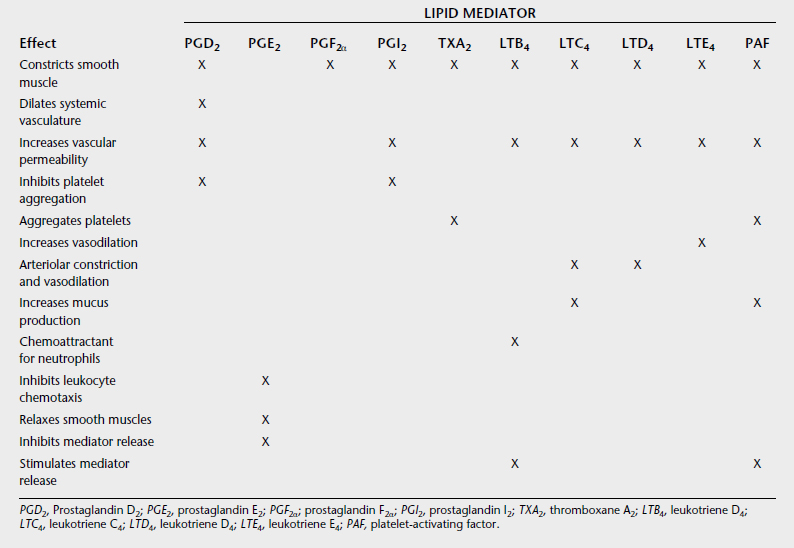
Prostanoids are lipid mediators that regulate the inflammatory response.14,15 The prostanoids group includes the prostaglandins (PGs), leukotrienes (LTs), and prostacyclin (PGI2), and they are the product of cyclooxygenase cleavage of arachidonic acid followed by endoperoxidation (Figure 1-2). The major sources of prostanoids in acute inflammation are the phagocytes, endothelial cells, and platelets. Prostanoids, in general, mediate the cardinal effects of pain, fever, and edema characteristic of the acute inflammatory response, but their particular roles are somewhat confounding and can be either pro- or anti-inflammatory (Table 1-2).16 Prostanoid production depends on the activity of the two isoforms of the cyclooxygenase enzymes within cells: COX-1, which is present in most cells and its expression is generally constitutive, and COX-2, whose expression is low or undetectable in most cells, but its expression increases dramatically upon stimulation, particularly in cells of the immune system. Increased COX-2 expression by inflammatory stimuli likely accounts for the high levels of prostanoids found in chronic inflammatory lesions and is the basis for the development of COX-2–specific inhibitors for treating chronic inflammatory diseases.17 However, studies using mice have indicated that the earliest prostanoid response to deleterious environmental stimuli depends on COX-1, and only as the inflammatory process progresses does COX-2 become the major source of prostanoids.18 Further, evidence for increased cardiovascular risk associated with COX-2 inhibitors has called into question the use of COX-2 inhibitors as treatments for inflammatory diseases.19,20
Both COX isoforms produce PGH2, which is the common substrate for a series of specific synthase enzymes that produce PGD2, PGE2, PGF2, PGI2, and TXA2 (see Figure 1-2). It is the differential expression of these enzymes within cells present at sites of inflammation that will determine the profile of prostanoid production. For example, mast cells predominantly generate PGD2, whereas resting macrophages produce TXA2 in excess of PGE2, although this ratio changes to favor PGE2 production after activation. Likewise, the biological effect of a prostanoid depends on its binding to G protein–coupled cell surface receptors. The receptors for PGF2, PGI2, and TXA2 are called FP, IP, and TP, respectively. In contrast, PGD2 acts through two receptors, the DP receptor and the recently identified CRTh2 receptor, and there are four subtypes of receptors for PGE2, termed EP1–EP4. The prostanoid receptors themselves are coupled to various G protein–coupled intracellular signaling pathways. The DP, EP2, EP4, IP, and one isoform of the EP3 receptor can couple to Gs and thus increase intracellular cAMP concentration, which in T cells and other inflammatory cells is generally associated with inhibition of effector cell functions. By contrast, the EP1, FP, IP, and TP receptors, as well as other EP3 isoforms, couple to Gq, and activation of these receptors leads to increased intracellular calcium levels and immune cell activation. Finally, TP, CRTh2, and yet another EP3 receptor isoform can each couple to Gi, causing cAMP levels to decline while also mobilizing intracellular calcium. Many cells of the immune system express multiple receptors that couple to these apparently opposing pathways. The impact of prostanoids present during an inflammatory response is thus determined by the array of receptors the cells express and the intracellular pathways to which they are coupled. Activation of these receptors, even when coupled to similar pathways, might evoke different responses because of differences in the levels of expression (both constitutive and induced) or in the patterns of desensitization. The role of prostanoids in a given inflammatory response depends not only on the presence of the lipid mediators in the lesion but also on the receptor profile on immune cells and the biochemical signaling pathways of these receptors.18 Thus PGE2 is considered proinflammatory because it promotes vasodilation by activating cAMP-coupled EP2 receptors on vascular smooth muscle and increases vascular permeability indirectly by enhancing the release of histamine and other mediators from tissue leukocytes such as mast cells. PGE2 is also the prostanoid responsible for fever production. However, as inflammation progresses, PGE2 synthesis by macrophages is enhanced as a result of increased expression of COX-2 and PGE-synthase, and the resulting increased levels of PGE2 inhibit leukocyte activation, mast cell degranulation, and relax smooth muscle contractions. In the lung PGE2 promotes bronchodilation through activation of Gs-coupled EP2 and EP4 receptors. Thus in these situations PGE2 may be considered anti-inflammatory.
Chemotaxis and Leukocyte Trafficking
One of the initial and most crucial aspects of the acute inflammatory response is the recruitment of leukocytes (primarily neutrophils) to the site of injury. Neutrophils constitute the first line of the cellular defense and are the initial cells involved in an inflammatory response. These phagocytic cells are derived from multipotent stem cells located chiefly in the bone marrow. Under the influence of a variety of signals provided from both within and outside of the bone marrow, these stem cells become committed to developing into cells of the granulocyte lineage. The critical signal is provided by a family of growth factors known as colony-stimulating factors (CSFs) that provide both proliferative and differentiative signals leading to the development of granulocytes and other leukocytes. Once released into the circulation, these cells must find their way to the site of the inflammatory response. The production of various chemotactic factors by host cells, bacteria, and other invaders causes various leukocytes to enter the circulation and be carried to the site of the injury.21 Chemokines are soluble proteins produced by host cells that induce the directional migration and activation of leukocytes, as well as other somatic cell types, and thus play a major role in the inflammatory response.22 Interleukin 8 (IL-8) plays a central role in this process. Other chemokines promote humoral and cell-mediated immune reactions; regulate cell adhesion, angiogenesis, leukocyte trafficking, and homing; and contribute to lymphopoiesis and hematopoiesis.23
The specific trafficking of leukocytes from the blood to inflammatory sites depends on both the production of chemotactic factors and the interaction of specific receptors on the leukocytes with corresponding adhesion molecules on the endothelial surface of the blood vessels. Neutrophil adherence is a two-step process first involving endothelial cell surface molecules known as selectins.9 Small venular endothelium overlying a site of inflammation and exposed to thrombin, platelet activating factor (PAF), IL1, histamine, or other mediators released by clotting, platelet activation, or mast-cell activation express P-selectin.24 P-selectin mediates the process in which neutrophils initially interact with the endothelial surface in a process known as “rolling,” in which the circulating neutrophil interacts with the endothelial cell before the actual adherence.25 Selectins function by binding to carbohydrate ligands present on the cell surface. In the case of neutrophils, the ligand is sialylated Lewis-X antigen for the endothelial E-selectin. The second part of the adherence process is the tight binding of integrins on the neutrophil surface with intracellular adhesion molecules (ICAMs) on the endothelial cell surface. Leukocyte integrins are heterodimeric proteins with distinct α and shared β polypeptide chains. The α and β chains can combine in different heterodimers to form multiple shared and unique specificities. Neutrophil expression of αMβ2 and αXβ2 is activation dependent. Neutrophils can be activated by a number of soluble proteins, including formylmethionylleucylphenylalanine (fMLP), N-formulated peptides present in bacterial but not eukaryotic proteins. Host factors present at the site of inflammation can also activate neutrophils, notably the complement proteins (C5a, C3a) and cytokines such as IL-8 and tumor necrosis factor (TNF), and immune complexes.26 Expression of integrins by activated neutrophils allows them to become tethered to the endothelial surface. The migration of neutrophils through the vascular wall is less well understood than these initial events leading to firm adhesion. The β2 integrins, as well as αvβ3, PECAM-1 and integrin-associated protein (IAP), appear to play a role in this process. Endothelial cell–produced IL-8 also is believed to have a critical role in this process. Once through the endothelium, the phagocytes will follow chemotactic signals and migrate toward the point of injury. They may adhere to other cells during migration to the site of inflammation, and these interactions also depend on αMβ2 and αXβ2 integrins. Migration through the extracellular matrix is mediated by β1, β3, and β5 integrins recognizing specific protein ligands.
Neutrophils recruited and activated in this manner will actively phagocytose microscopic invaders and attempt to destroy them using reactive oxygen products generated via an NADPH-oxidase–dependent “respiratory burst.”25,27 In the process the neutrophils release additional pro-inflammatory mediators, thus amplifying this response. Among those cells attracted to the area are natural killer (NK) cells capable of lysing virus-infected and other abnormal cells. The production of interferon-α/β by macrophages and other cells enhances the cytolytic activity of the NK cells. The NK cells themselves can be the source of interferon-γ, another pro-inflammatory cytokine. Depending on the magnitude of the initial insult and the susceptibility of the invader to neutophil-mediated destruction, the inflammatory response may be either acute or chronic.
Acute inflammation is thus a rapid response to an injury that is characterized by accumulations of fluid, plasma proteins, and neutrophils that rapidly resolves once the initial inflammatory stimulus is removed. Deactivation signals include PGE2, cortisol, IL-10, and transforming growth factor-β (TGF-β). Some of those chemotactic agents responsible for initiating the response (IL-8, FMLP, C5a, LTB4, and PAF) also serve to downregulate its intensity by inducing the shedding of IL-1 receptors from neutrophils.28 The shedding of this decoy receptor may have anti-inflammatory effects as it effectively binds and neutralizes this cytokine. Likewise, many of the acute phase proteins are thought to have immunomodulatory activity downregulating neutrophil function.29 Acute inflammatory responses may often be subclinical and resolve without complications. However, if the invader is resistant to neutrophil-mediated destruction or the degree of injury is large, the response may become more chronic with the added recruitment of macrophages and lymphocytes, and fibroblast growth.
The essential characteristic of the innate immune response is that it does not exhibit specificity for the invading organism. Thus the induction of an innate immune response does not require prior exposure to the invading organism nor is it augmented by repeated exposure to the same organism. Whereas resistance may be genetically controlled, the genes encoding resistance are not found within the gene complex that controls adaptive immune responses. In most instances these mechanisms are adequate for eliminating casual invaders. However, pathogenic organisms have evolved various methods for avoiding elimination. In response to these organisms, the specialized cells and products of the adaptive immune response are mobilized.
ADAPTIVE IMMUNITY
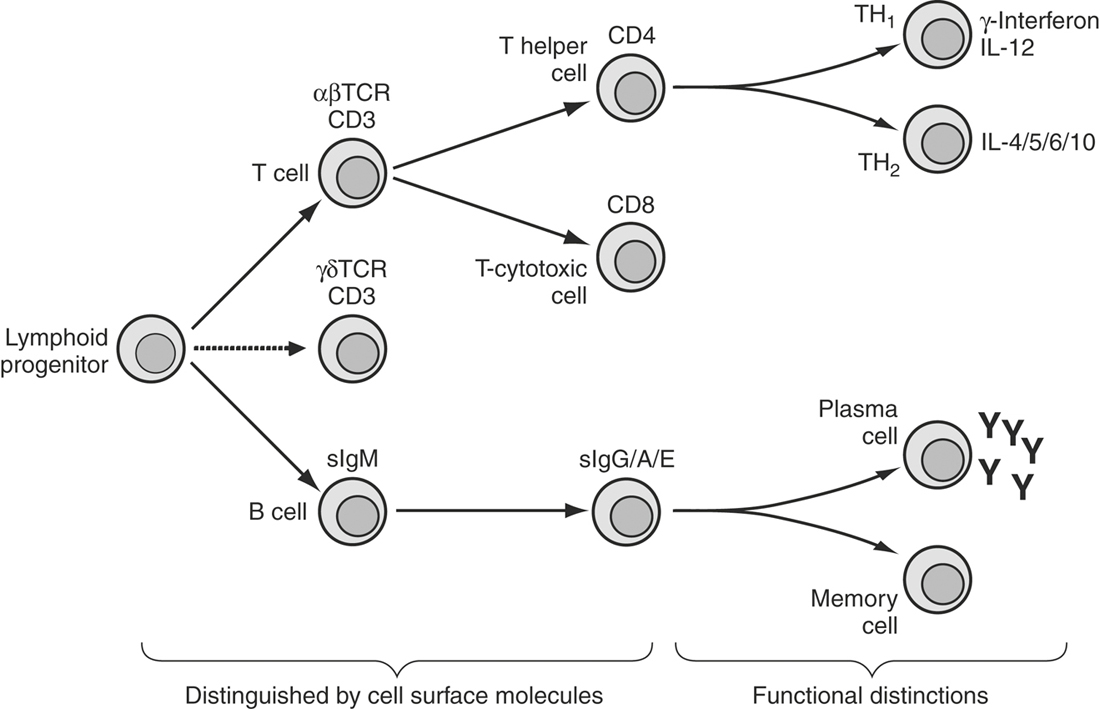
The adaptive immune response is initiated in response to an encounter with a foreign agent and depends on antigen-specific immune responses mediated by different divisions of the lymphocyte family (Figure 1-3). In contrast to the nonspecific nature of the innate immune response, an important characteristic of the adaptive immune response is the specificity of this interaction. Thus exposure of the host to a particular microbe or parasite results in the induction of immune responses that are directed against specific components of the invading organism that do not affect unrelated organisms. The specificity of the adaptive immune response is the result of the interaction of specific molecular structures or antigens of the invader with antigen-specific receptors on lymphocytes. All types of chemical structures can serve as antigens, but not all antigens can induce an immune response. Immunogens, those antigens that can stimulate an immune response, are usually high molecular–weight, chemically complex molecules. Proteins, nucleic acids, lipids, and polysaccharides can all serve as immunogens. Large immunogens, such as proteins, contain multiple antigenic determinants or epitopes, which interact with lymphocytes via their antigen-specific receptors. Haptens consist of single antigenic determinants and can effectively combine with the binding site of antibody molecules. However, because they consist only of a single antigenic determinant, they cannot cross link B-cell receptors (antibody molecules), and they are also unable to stimulate T cell responses. Haptens therefore cannot stimulate an immune response unless multiple haptens are physically attached to a larger molecule, known as a carrier. Although these distinctions between antigens, haptens, and immunogens appear minor, they provide the underlying basis for our understanding of many allergic and autoimmune responses.
Although the induction of an antibody response requires the interaction of B and T lymphocytes, these cells recognize different epitopes on the same antigen. Indeed, antigen recognition by B cells and T cells is fundamentally quite different. B cells, and antibodies, recognize antigens in solution or on cell surfaces in their native conformation, whereas T cells recognize antigen only in association with self molecules known as major histocompatability complex antigens found on most cells’ surfaces. The adaptive immune response thus differs from innate immunity in that it is antigen driven and those cells that mediate the adaptive immune responses, T and B lymphocytes, express specific receptors for the antigen. Because the immune system will respond to the antigens of both live and killed pathogens, it is possible to stimulate immunity without causing infection, which is the basis of vaccination. Although this principle appears to be straightforward, vaccination does not always yield the expected result. Why some vaccines work and others fail is a complex issue, a major component of which is the nature of the antigen-specific receptors of lymphocytes.
Immunoglobulin: Antigen-specific Receptor of B Lymphocytes
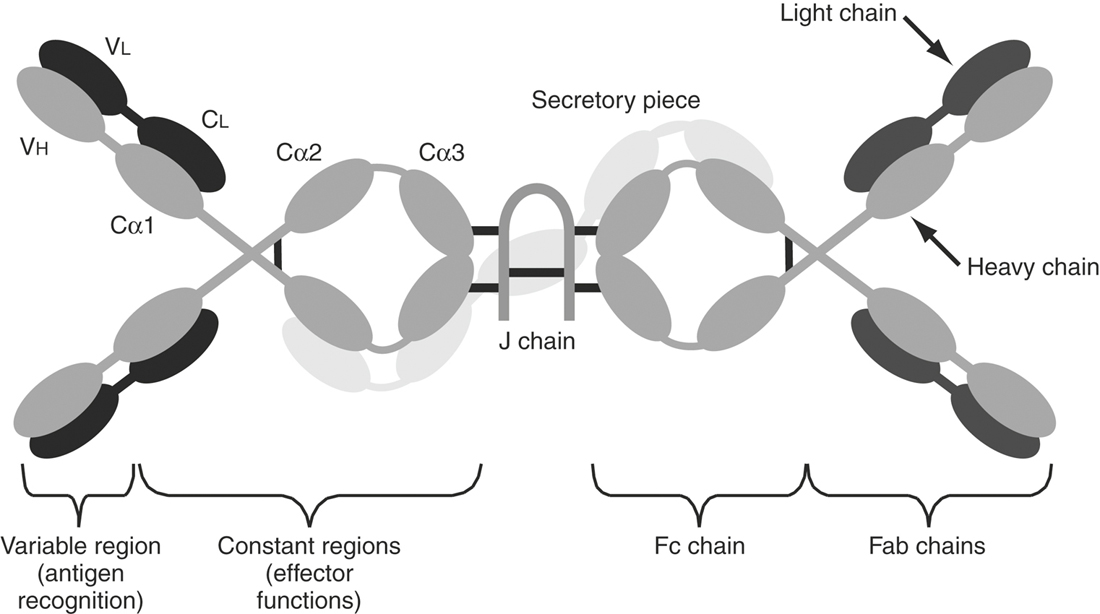
The antigen-specific receptor of the B cell is cell surface–bound antibody. An antibody molecule is composed of two identical light chains and two identical heavy chains that form a disulfide-linked Y-shaped molecule (Figure 1-4). The light chain can be divided into two domains, a conserved carboxy-terminal domain and a highly variable amino-terminal domain. Analysis of heavy chains reveals a similar domain structure, with the amino-terminal domain being highly variable and the presence of three constant domains. The antigen-binding region of an antibody molecule is formed by the association of the amino ends of a light and a heavy chain, whereas the carboxyl end of the heavy chain determines the isotype of the molecule. Five different isotypes, or classes, of antibody molecules have been identified in most species, including the horse: IgD, IgM, IgG, IgA, and IgE (Table 1-3).30,31 Additionally, the IgG isotype can be subdivided into subclasses based on physico-chemical properties. Analysis of equine genomic DNA has indicated the existence of one IgM, one IgD, one IgE, one IgA, and seven IgG genes.31 Before the availability of genetic characterization of the equine immunoglobulin heavy chain gene loci, at least four IgG subclasses were identified by physico-chemical means and defined serologically by monoclonal antibodies as IgGa, IgGb, IgGc, and IgG(T).32 Each of these “classical” (older) IgG subclasses has been identified as a gene product of one or more of the equine heavy chain gene loci.30,31 It appears that both IgGb and IgG(T) are encoded by two loci each, raising the possibility that these classical subclasses may each comprise two distinct subclasses. At the time of writing, this remains uncertain, although ongoing studies are starting to resolve the issue.30 The gene product of at least one IgG heavy chain locus is not defined by the classical IgG subclasses, and it remains to be determined whether it is expressed as a protein and what its role may be.31 Given the current remaining uncertainties as to the role of equine IgG subclasses as defined by nomenclature based on heavy chain gene locus, this edition of this text will continue to use the classical nomenclature.
TABLE 1-3 Immunoglobulin Isotypes
| Isotype | Immunologic Function |
|---|---|
| IgD | Antigen receptor of naïve B lymphocytes. An IgD heavy chain gene has been identified in the horse, and it is probably expressed. |
| IgM | Surface IgM is found on naïve, activated, and memory B cells. Secreted IgM is a pentamer and represents the major antibody produced during a primary response. IgM efficiently mediates agglutination, neutralization, opsonization, and complement activation. |
| IgG | The principle immunoglobulin found in plasma representing up to 80% of the total immunoglobulin concentration. Various subclasses of IgG have been identified (see text). The classical view is that there are four IgG subclasses in the horse (IgGa, IgGb, IgGc, and IgG(T)) defined by physicochemical properties and monoclonal antibodies, although seven IgG heavy chain genes have been identified and there is evidence that all are expressed. The major functions of IgG include opsonization and neutralization reactions. IgGa and IgGb are effective in fixing complement and participates in antibody-dependent cellular cytotoxicity (ADCC) while IgGc and IgG(T) are not, although they appear to play an important role in exotoxin neutralization and immunity to parasites. As our understanding of IgG heavy chain gene expression and function increases, a new nomenclature (i.e., IgG1–7) will replace the classical nomenclature (i.e., IgGa). |
| IgA | IgA, the most abundant antibody in secretions (e.g., tears, mucus, saliva, colostrum) is a dimer composed of two IgA molecules joined by a J chain. IgA in the plasma is predominantly monomeric. IgA antibodies can be neutralizing but only activate complement via the alternative pathway. |
| IgE | Most IgE is found associated with the surface of mast cells and basophils and only very small amounts are present in the plasma. The cross-linking of two IgE moleulces with specific antigen results in the degranulation of the mast cells and basophils. Thus IgE is the primary antibody responsible for Type I hypersensitivity reactions and appears to play a central role in immunity to parasites. |
Ig, immunoglobulin.
Data from Wagner B. Immunoglobulins and immunoglobulin genes of the horse. Dev Comp Immunol 2006;30:155-164; Lewis MJ, Wagner B, Woof JM. The different effector function capabilities of the seven equine IgG subclasses have implications for vaccine strategies. Mol Immunol 2008;45:818-827; and Wagner B, Miller DC, Lear TL, Antczak DF. The complete map of the Ig heavy chain constant gene region reveals evidence for seven IgG isotypes and for IgD in the horse. Journal of Immunology 2004;173:3230-3242.
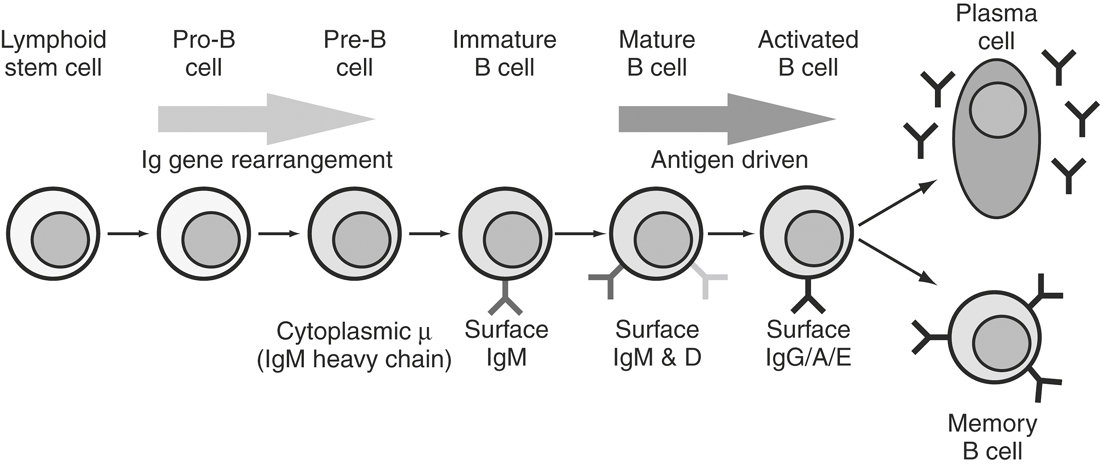
Membrane-bound IgM and IgD serve as the antigen-specific receptors for B lymphocytes. Each contains a membrane-spanning region near its carboxy end that is inserted into the mRNA during differential splicing of the heavy chain exons. Although rarely detectable in the circulation, IgD is present in large quantities on the surface of naïve B lymphocytes. Following activation, the surface expression of IgD is lost, although the cell may continue to express the membrane form of IgM. Early in an immune response, the B cell secretes large amounts of the pentameric form of IgM. As the immune response proceeds, the B cell will switch the isotype of its heavy chain. Isotype switching involves the substitution of one heavy chain–constant region in place of another. The genes encoding the five different constant regions of the heavy chain are sequentially arranged on the chromosome (Cδ, Cμ, Cγ, Cε, and Cα). Initially, the first two constant region genes encoding the δ and μ constant regions are used to form the heavy chain. The 5’ region of each constant region gene segment contains repetitive regions of DNA known as switch sequences.33 The switch sequences appear to play a role in this rearrangement and may serve as the target for specific recombinases. When switching occurs, a new constant region segment is selected and the intervening genes are removed either by splicing or looping out. Isotype switching affects only the heavy chain–constant domains and has no effect on the antigen specificity of the immunoglobulin molecule. The signal for B cells to undergo isotype switching is provided by T lymphocytes in the form of various cytokines.34 For example, IL-4 induces isotype switching to the IgE isotype, whereas interferon-γ blocks this induction and augments IgG production.35,36 IgA is produced in response to the combination of the cytokines IL-4, IL-5, and transforming growth factor-ß (TGF-β).37
The antigen specificity of a particular antibody molecule (and the B cell that produces it) is determined by the combination of the variable domains of the light and heavy chains. The association of these two domains results in the formation of an antigen-binding groove or pocket that contains regions of hypervariability that define the specificity of a particular antibody molecule. It has been estimated that more than 108 different antibody specificities are possible. The generation of this tremendous amount of diversity in antibody specificity occurs during B cell ontogeny in the bone marrow.38 Within a given B cell, the genes encoding the heavy and light chains of an antibody molecule are organized into specific gene segments. Thus the light chain is formed from variable (Vl), joining (Jl), and constant (Cl) gene segments that together form the variable and constant domains of the light chain. In the germ line of an undifferentiated cell, several hundred different Vl and several dozen Jl gene segments can be found. Likewise, the heavy chain of a B lymphocyte is composed of VH, diversity (D), and JH segments that form the variable domain, and these join to the constant region genes to form the complete heavy chain molecule. Similarly, in the germ line a large number of VH gene segments and a smaller number of D and JH segments are found. During the differentiation of a B cell (Figure 1-5), there is the sequential selection and rearrangement of a VL segment with a JL segment and the accompanying deletion of intervening VL and JL segments (Figure 1-6). The rearranged VJC sequence is then transcribed into mRNA and translated into the light chain. A somewhat similar sequence follows for heavy chains except that two rearrangements are necessary, a D to JH rearrangement followed by a VH to DJH rearrangement. Once completed, the VDJ segment is brought into the proximity of the appropriate CH segment and transcribed. Not all of the gene segment rearrangements produce functional genes. Because a B cell has two sets of heavy chain genes, one on each chromosome, and most species, including the horse, have two different sets of light chain genes,39,40 there are several chances to form appropriate heavy and light chains. Once the heavy and light chain gene segments are successfully recombined, the genes on the sister chromosome neither recombine nor are they expressed. This process of allelic exclusion ensures that the B cell produces antibodies of a single specificity. Although this random assortment of gene segments accounts for much of the diversity in antibody specificity, additional mechanisms are also involved, including junctional diversity, which results from the imprecise joining of gene segments and somatic mutations. Somatic mutations are point mutations in the hypervariable region of either the heavy or light chain that occur during the proliferation of antigen-activated B lymphocytes. Such mutations appear to play a role in increasing antibody affinity for its antigen. Thus fewer than 1000 genes can give rise to more than 108 molecules of the various specificities needed to recognize the vast number of antigens the host may encounter.
TcR and CD3 Complex: Antigen-specific Receptor of T Cells
T lymphocytes can be differentiated from B lymphocytes in that they do not express surface immunoglobulins but instead express the T cell receptor (TcR). T cells also express another antigen called CD3. (The designation CD stands for cluster designate and is the result of an international workshop to standardize the terminology used to describe leukocyte surface antigens recognized by monoclonal antibodies.) The TcR and CD3 form a multimeric complex on the T cell surface, and this complex is involved in antigen-specific recognition.41 The TcR structure was first identified using antibodies that recognized a surface antigen expressed on a cloned T lymphoma cell line. This antibody recognized a disulfide-linked heterodimer composed of an acidic (α) and a basic (β) protein of 40 to 45,000 molecular weight. Similar heterodimers were found on a variety of antigen-specific T cell lines but not on B cells. Peptide mapping studies of the α and ß chains from many different T cell lines demonstrated that they contained variable and constant domains reminiscent of immunoglobulin structure. Further analysis indicated that, like immunoglobulin genes, the TcR genes underwent gene rearrangements during T cell development. Subsequently, two additional TcR genes were identified, the γ chain and δ genes corresponding to a second heterodimer. Thus two TcR exist, an α/β heterodimer that constitutes the TcR on almost 90% of all T cells and a γ/δ heterodimer present on approximately 10% of the peripheral T cells. The significance of these two different TcR heterodimers has not yet been determined. It should be noted that γ/δ T cells have not yet been identified in the horse. It is clear that γ/δ T cells represent a functionally distinct population of T cells typically associated with mucosal surfaces.42 As such they are thought to play an important role in immunologic surveillance.
Analysis of the predicted amino acid sequences for the TcR proteins confirmed a structural similarity with antibody molecules. One peculiarity in the structure of the TcR was observed from the amino acid sequence analysis. Whereas both the α and β chains of the TcR contained a transmembrane region, both proteins had very short cytoplasmic tails. It therefore seemed unlikely that TcR itself could transmit any cytoplasmic signal in response to antigen binding. This led to the search for other proteins associated with the TcR. Solubilization of the T cell membranes revealed that five other proteins could be immunoprecipitated with the TcR. Similar results were obtained when anti-CD3 antibodies were used. Thus the TcR heterodimer is noncovalently associated with the CD3 complex of proteins. The five proteins of the CD3 complex (γ, δ, ε, ζ, and ξ) are involved in signal transduction following TcR binding to antigen.43 Unlike the TcR α and β proteins, the CD3 proteins have large intracellular domains, some of which are phosphorylated in response to stimulation of the TcR. In addition to providing a signaling mechanism for the TcR, the CD3 complex is also required for the expression of the TcR heterodimer on the cell surface.41
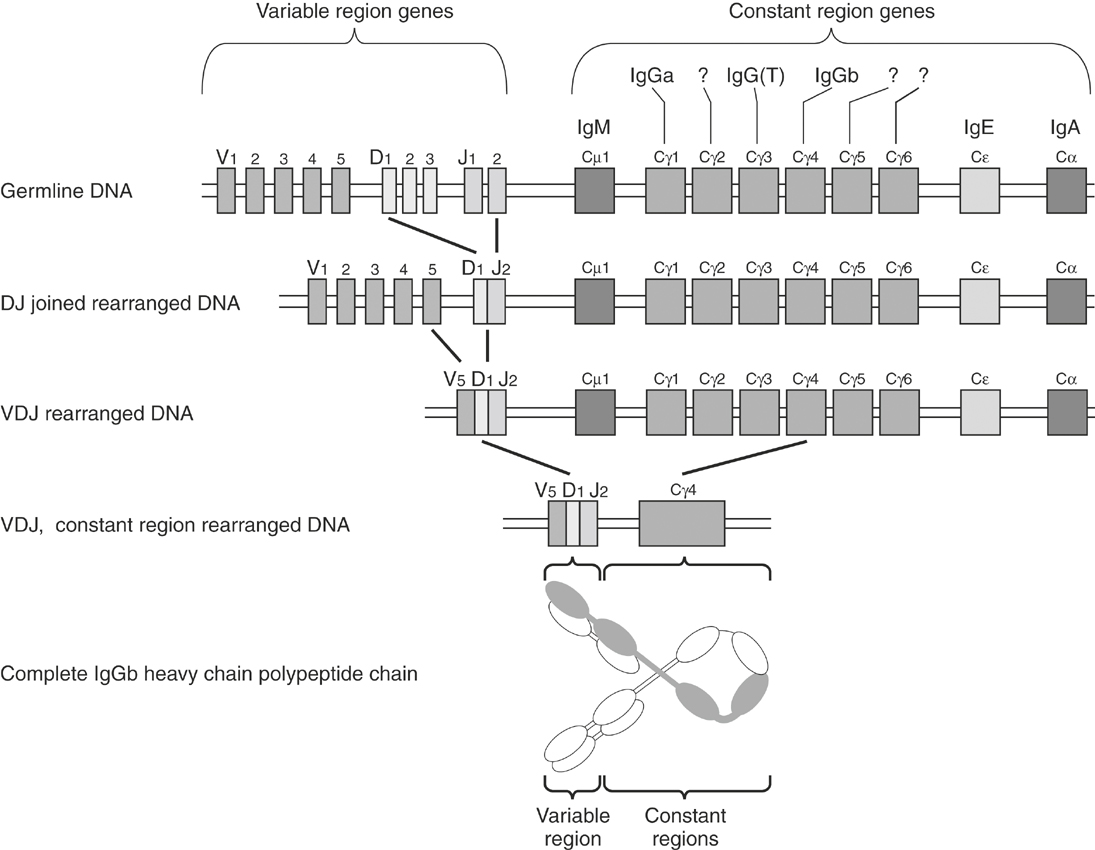
 to the known equine heavy chain constant region gene loci. In the first step in somatic recombination a D and a J gene segment are joined, and in the second step a V gene segment is joined to complete the VDJ recombination and form a gene capable of encoding the variable region. Subsequently one of the seven equine γ heavy chain constant regions, labeled with their corresponding IgG subclass when known, was selected to complete the gene rearrangement. Because the Cγ4 heavy chain constant region gene was selected, this leads to production of an IgGb heavy chain.
to the known equine heavy chain constant region gene loci. In the first step in somatic recombination a D and a J gene segment are joined, and in the second step a V gene segment is joined to complete the VDJ recombination and form a gene capable of encoding the variable region. Subsequently one of the seven equine γ heavy chain constant regions, labeled with their corresponding IgG subclass when known, was selected to complete the gene rearrangement. Because the Cγ4 heavy chain constant region gene was selected, this leads to production of an IgGb heavy chain.


