CHAPTER 38 Salmonellosis
ETIOLOGY
Salmonellosis is disease caused by an enteric or systemic infection with a bacterium of the genus Salmonella. This genus contains two species, Salmonella enterica and Salmonella bongori. To the authors’ knowledge, S. bongori has never been reported as the cause of equine salmonellosis. S. enterica includes six subspecies with more than 2000 serovars.1 All these species and subspecies are potentially pathogenic, but the virulence of many is undefined; the vast majority of clinical cases are associated with a single subspecies, S. enterica subsp. enterica. The National Veterinary Services Laboratory (NVSL) annually provides a valuable summary report of the frequency of clinical and nonclinical serovars by host species of origin; these data show that relatively few serovars of S. enterica subsp. enterica consistently account for a large percentage of clinical isolates.2–6
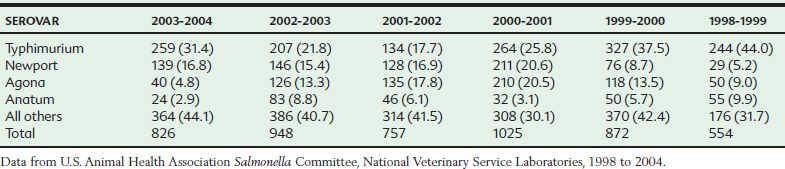
In horses the most frequently occurring serovars isolated from clinical cases vary somewhat from year to year, but Salmonella Typhimurium, Newport, Anatum, and Agona are consistently among the most frequent isolates from equine salmonellosis cases in the United States. The relative ranking of these common serovars undergoes periodic shifts as epidemic strains emerge and disappear. For example, in the mid 1990s, Salmonella Typhimurium was relatively predominant, reflecting the global epidemic of the pentaresistant Typhimurium strain DT104. During the past 5 years (as of 2006), however, Salmonella Newport has increased in relative frequency, with the emergence of the North American cmy-2 cephalosporinase-producing strains (Table 38-1).2–6
Table 38-1 Frequently Occurring Equine Salmonellosis Serotypes: Number (Percent) of All Equine Serotypes
Certain different Salmonella serovars produce distinct clinical syndromes that result in their classification as “host adapted” or “non–host adapted.” Serovars with strong host specificity and with clinical similarity to human Salmonella typhi infections are considered host adapted, whereas non– host-adapted serovars show minimal host specificity and a different clinical presentation. Host-adapted serovars produce systemic infections characterized by bacteremia, fever, and systemic signs. In host-adapted salmonellosis, diarrhea is not observed or, if present, is a relatively minor component.7 The equine host-adapted serovar, Salmonella Abortus-equi, causes a disease with bacteremia and infectious abortion as its principal manifestations.8 In contrast, non–host-adapted serovars typically produce localized infections of the intestine and colon with enterocolitis and diarrhea as the predominant components.7 Salmonella Abortus-equi is common in many tropical countries but does not occur in the United States; there have been no reports since at least 1978 (K.E. Ferris, personal communication). Where Salmonella Abortus-equi is not present, the principal manifestation of equine salmonellosis (the United States at present) is enterocolitis. Nevertheless, even non–host-adapted serovars can produce septicemic disease (although generally accompanied by enterocolitis) in highly susceptible hosts such as neonates.
For some purposes, strain-specific identification of Salmonella isolates within serovars is useful to identify potential nosocomial and other common-source outbreak strains and to investigate the epidemiology of Salmonella infection. Salmonella strain typing can be accomplished by different methods, including phenotypic procedures such as phage typing and antimicrobial resistance typing.9 Phage typing is a highly reproducible and discriminatory method, but this procedure is not widely available.10 More recently, genetic typing with pulsed-field gel electrophoresis (PFGE) after chromosomal digestion with infrequent-cutting restriction endonucleases has been increasingly used as a means of identifying strain types of Salmonella infection.11 Both phage typing and PFGE typing may be insufficiently discriminatory to identify subtypes within disseminated clonal types of Salmonella spp. For example, isolates of the epidemic type Salmonella Typhimurium DT (phage type) 104 obtained from widely distant sites around the world are frequently homogenous by these tests. Even more discriminatory tests, such as analysis of variable-number tandem repeat (VNTR) deoxyribonucleic acid (DNA) sequences, have proved useful to differentiate epidemiologically linked isolates of highly conserved strains such as DT104.12
EPIDEMIOLOGY
Source of Infection
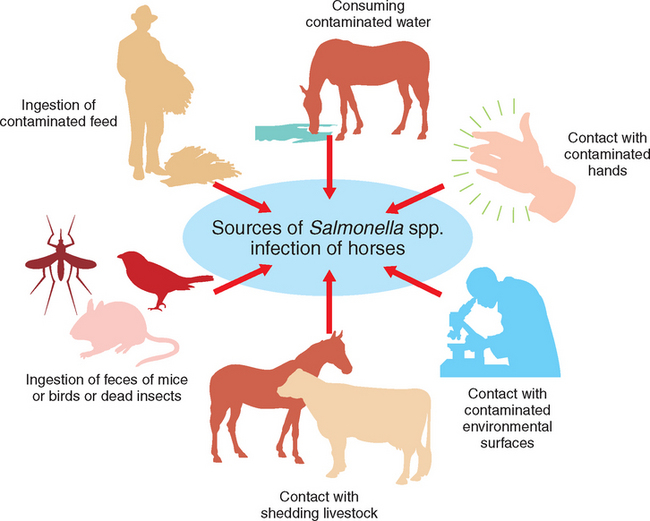
The primary route of exposure is likely oral intake of the organism; thus there are multiple potential sources of infection for equids (Fig. 38-1). These include consumption of contaminated water or feed and oral contact with the feces of infected animals, with contaminated environmental surfaces, or with animal care worker’s contaminated hands or equipment. It is unknown what percentage of Salmonella infections in equids are caused by these various potential sources. Aerosol exposure to the agent has been proposed in other animal species.30,31 Although evidence of this route of exposure in horses is lacking, it theoretically could occur.
The most frequently reported outbreaks of equine salmonellosis have been among hospitalized patients. At least some of these outbreaks are associated with nosocomial spread of a given serotype of Salmonella spp. The source of the organism in these outbreaks is often not definitively determined. Multiple potential sources that culture positive for the agent, such as the equipment or hands of health care workers or environmental surfaces, are identified through outbreak investigations.32,33 However, which of these potential sources play the major role in a specific nosocomial infection is not always determined, and a different source may be involved for various patients during an outbreak.
Clinically normal horses shedding Salmonella in their feces potentially pose a risk to animals with which they have direct contact and can contaminate the environment, which serves as a source of the organism to susceptible animals.34 A larger percentage of horses appear to shed the organism in summer than in winter months.35–37 Other livestock could also be a source of Salmonella for horses. Through surveillance of all large animal hospital patients, cattle (specifically dairy cattle) have been found to shed Salmonella spp., more than equids or other large animal patients.38
Salmonellae can persist in the environment in fecal matter for months to years depending on the serotype, moisture content, and temperature conditions.39,40 In one study, Salmonella Dublin persisted in dried bovine feces for up to 5 years.41 In another study, Salmonella Cholerasuis was more persistent when feces were in the dried (13 months) versus wet form (3 months).39 In veterinary hospitals, where most outbreaks of equine salmonellosis have been described, environmental contamination with potential persistence of the organism may be a source of infection for susceptible animals, as are fomites and contaminated hands of hospital workers.
Feed is a potential source of Salmonella for horses. Most commercial equine feeds are manufactured using good manufacturing procedures (GMP), but none is certified to be Salmonella free. A National Animal Health Monitoring System (NAHMS) survey estimated that 0.4% of grain or concentrate that was the primary feed source for horses was positive for Salmonella spp., and that the serotypes detected were not those typically associated with clinical disease in horses.37 Thus it would appear from this study that concentrate source is rarely positive for Salmonella spp., on most equine operations. However, contaminated feed given to broodmares who then shed the organism in their feces was implicated as a cause of an outbreak of salmonellosis among neonatal foals in California. The serotype implicated was Salmonella Ohio, and biochemical profiles, restriction endonuclease analysis, and ribotyping demonstrated that feed and poultry isolates, most of which were in geographic proximity to the broodmare farm, were indistinguishable from the outbreak isolates.42 One report identified maize silage as containing Salmonella Typhimurium that resulted in fatal diarrhea and colic in horses to which it was fed.43 Pelleting likely decreases the number of salmonellae in contaminated feed, depending on the temperature used in the processing procedure.44
Apparently, components of livestock feed could be contaminated with Salmonella spp. In September 1990 the Food and Drug Administration Center for Veterinary Medicine (FDA-CVM) announced a goal of having zero salmonella contamination of animal feed ingredients and finished feed. One step in the process toward this goal was for FDA to perform a monitoring program of animal feeds. FDA conducted a survey in 1993 of processors manufacturing either animal or vegetable protein products used in animal feeds to determine the prevalence of Salmonella spp.45 The findings of the study represented 4530 individual samples. Salmonella was detected in 56.4% of animal protein samples and 36% of the vegetable protein products. In an FDA report, 34% of animal protein– derived feed samples were positive for salmonella in 2002 and represented 27 serotypes.46 In a 2-year study of Salmonella Typhimurium DT 104 on cattle farms, feed and grain sources remained contaminated throughout the study.47 An evaluation of inoculated swine feed and meal found a difference in survival of various salmonellae serotypes.48 For example, Salmonella Anatum persisted for 429 days in ground feed and 299 days in meat and bone meal, Salmonella Infantis was present at 723 days in feed and at 588 days in meal, and Salmonella Enteritidis survived approximately the same amount of time in feed and meal (728 days in feed and 750 days in meal).48 It was speculated that these non–host-adapted strains survived longer than host-adapted types evaluated, such as Salmonella Typhisuis and Cholerasuis.48
Equine feed sources such as grain or other concentrate sources also could become contaminated after processing once at the equine premises. Potential sources of contamination include the feces of rodents and birds or bodies of insects. Rodents have been proposed as a reservoir for Salmonella spp.,44,49 and have been determined to carry the same strain as was involved in an equine salmonellosis outbreak.32 Insects such as flies can become contaminated with Salmonella spp.50 when they contact infected feces or surfaces and then carry Salmonella to susceptible animals, which may either ingest the insect when eating or the insect may come into contact with the horse’s oral surfaces.
Pastures could become contaminated with Salmonella spp., if organic fertilizers or bone meal are used, if runoff occurs from surrounding animal holding facilities, or if contaminated water supply is used for irrigation or sprinkling. Feces passed by infected animals being housed on the pasture or from wildlife that may have access to the pasture (e.g., birds, wild mammals) could contaminate a pasture. Soils have been reported to remain positive for Salmonella spp., for a variable period from 120 to 280 days.51 Also, hay sources could become contaminated by these same means and then, when harvested, could act as a source of Salmonella to susceptible horses.
The role that contaminated water sources play in equine salmonellosis is unknown. Surface water (e.g., creeks, irrigation ditches, ponds or lakes) could be contaminated with the organism. In a national study of equine health and management, 10% to 33% of equine operations used surface water as the primary water source for equids.52 No testing of these water sources was performed as part of this study. Salmonella survives in pond water for 115 days.51 This likely varies with the serotype of Salmonella and environmental temperature, as well as other characteristics of the water (e.g., pH, salinity). Freezing reduces the total number of organisms, but survivors may remain viable and infective for months.51
Prevalence
The definition of salmonellosis includes infection with the organism as well as the occurrence of detectable disease signs secondary to infection,53 although some reports on equine salmonellosis also include animals shedding the organism without signs of disease.34,35 This is likely in part a result of the transition from early investigations focusing only on animals with clinical disease to the current interest in surveillance of the general population or overall hospital population for shedding of the organism. Therefore, all these factors need to be considered when describing Salmonella infection or shedding by equids. It has been proposed that three types of salmonella-infected horses may exist: carrier without fecal shedding, carrier with fecal shedding, and shedder with clinical disease.35 Also, there could be a “pass-through phenomenon” in which the horse is not infected (carrier) but instead ingests the organism and passes it through the gastrointestinal (GI) tract without any recognition of the agent’s presence (no serologic or local GI response to the organism).
The prevalence of clinical salmonellosis in the general equine population is unknown. During outbreaks of salmonellosis on a given farm or in a given veterinary hospital, the attack rate is variable, and there are few reports of the actual prevalence of clinical cases; instead, the number of cases is reported, but the number of animals at risk is usually not provided. In the hospital setting this may be the case because by the time the outbreak is recognized, the number of patients or patient days at risk is not available retrospectively. The prevalence of Salmonella infection as a cause of disease among horses with diarrhea has been evaluated in a Dutch study.54 During 1990 and 1991, 380 fecal samples were collected from horses that were referred for treatment of diarrhea. Most horses had a single fecal sample collected from the rectum, or if they died soon after arrival, samples were collected at necropsy. Of these samples, 18% (69/380) were positive for Salmonella spp. The most common serotype identified was Salmonella Typhimurium (43/69). Other serotypes identified less frequently included Salmonella Hadar (3/69), Arizona (2), Enteritidis (2), Virchow (1), Blockley (1), and Bareilly (1).
The prevalence of fecal shedding in a national study of the general horse population sampled while on their home premises, based on a single sample per animal, was 0.8%. The prevalence of fecal shedding by horses in this study was higher in the summer months (1.1%) than in the winter months (0.2%) and in the southern region (1.4%) than in the northern region (0.2%) of the United States.37 A total of 16 different serotypes were identified, with the most common serotype being Muenchen, followed by Newport, Schwartzengrund, and Typhimurium.
One of the first studies of the prevalence of fecal shedding among the general hospitalized equine population was conducted as a surveillance program at a university teaching hospital in the late 1970s. A total of 1451 horses were sampled, with a reported 3.2% prevalence of fecal shedding of Salmonella.35 The serotypes identified included Typhimurium, Typhimurium var Copenhagan, Infantis, Montevideo, Meleagridis, and Drypool, as well as untypeable isolates. Seasonal variability was marked in detection of fecal shedding, with the highest incidence in the late summer and early fall and the lowest in the spring. Of the 46 horses shedding Salmonella in this study, 18 had diarrhea, and seven deaths were attributed to salmonellosis. Since this early report, many more reports on the prevalence of shedding Salmonella by hospitalized equids have been published. In a study of the general equine hospitalized population (anticipated hospital stay ≥3 days, 246 horses), the prevalence of fecal shedding was 7%, with serotypes identified including Oranienberg, Newport, and Arizona, in descending order of frequency, followed by Newington, Drypool, Anatum, Thompson, and Meleagridis, each represented by single isolates.55 Only 3 of the 18 culture-positive horses in this study were admitted for diarrhea.
Some reports on the prevalence of fecal shedding of Salmonella spp., were based on a subset of the hospital population, for example, patients admitted for colic or needing intensive care. The prevalence of fecal shedding among colic patients (246 horses, with an average of three samples per horse) at a veterinary teaching hospital was 9%.56 The serotypes were Typhimurium, Infantis, Muenchin, and Anatum, in descending order of frequency, followed by a similar number with Oranienburg, Montevideo, and Thompson, all with the same frequency. In a second study of colic patients (based on culture of feces or rectal swab samples) the prevalence was 13% (100 horses, with an attempt to collect five samples per horse, unless it died before day 5), and the most common serotype identified was Senftenberg, followed by Typhimurium, then London and Agona.57 Senftenberg was also the most frequently isolated serotype from horses “without colic” in this report. In a survey of equids admitted to a veterinary teaching hospital intensive care unit over a 4-year period (1583 horses, with daily collection of fecal samples for culture), the overall prevalence of shedding was 5.5%, and the most common serotypes identified were Typhimurium and Krefeld; other types identified included Anatum, Agona, Enteriditis, Heidelberg, Muenster, Newington, Oranienberg, Poona, and Tennessee.58
The prevalence of carrier horses that are not shedding the organism in their feces as detectable by culture is difficult to determine. There are few studies of long-term, repeated fecal culture and sampling of mesenteric lymph nodes and other sites that Salmonella spp. may reside. In one study of 85 equids undergoing necropsy for various causes other than salmonellosis in England, 20% had Salmonella spp. isolated from one or more sites, with the mucosa of the cecum and large colon being the most common sites harboring the organism. No details on culture method or sampling method were included in this report.59 A culture survey at one equine slaughter plant showed a 70% prevalence of carrier horses.60 In a second slaughter plant study the prevalence of isolation was 27%.61 In a study of 102 horses that underwent necropsy between April and December 1994 at a veterinary teaching hospital, only two foals had Salmonella spp., recovered from the mesenteric lymph nodes.62 These authors concluded that the results of cross-sectional studies using culture to determine Salmonella infection should be interpreted with caution because the results of prevalence from a single facility may not reflect the prevalence of infection in the general population. In one of the slaughter plant surveys the authors speculated that the horses were becoming infected while at the slaughter plant because the serotype of Salmonella identified was similar to that obtained from birds sampled from corrals where the horses were housed before slaughter.63
The prevalence of fecal shedding by horses as determined by a PCR test can be quite different from prevalence determined by culture. It is important to recognize that PCR results vary depending on which primers are used,64 so when describing the prevalence of shedding of Salmonella by horses, it is important to distinguish if prevalence for shedding was detected by bacterial culture or by PCR test as well as what primers were used in the PCR test. For example, 71 of 110 horses tested positive using a PCR test with the hisJ gene on feces, whereas only 11 of these same horses were positive based on fecal culture.65 From this example, therefore, the prevalence of fecal shedding based on PCR would obviously be much higher than that based on culture of fecal samples. At this time, most prevalence data reported for shedding are based on culture rather than PCR results.
The prevalence of serotypes identified among Salmonella isolates of equine origin is reported annually at the U.S. Animal Health Association Salmonella Committee meeting by the NVSL staff2–6 (see Etiology for annual statistics).
Risk Factors
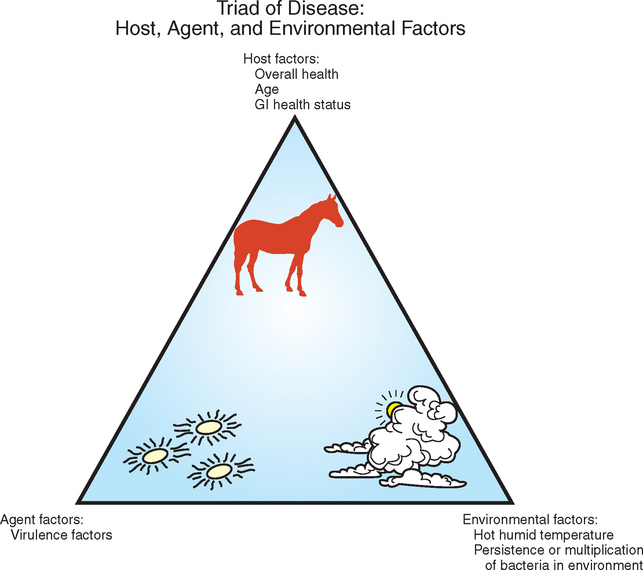
The ability to infect horses with Salmonella and the subsequent development of clinical disease depend on multiple factors (Fig. 38-2). Age may be a risk factor for development of clinical illness, with foals more likely to develop disease than adults.79,80 Stress may be a predisposing factor for initiating equine salmonellosis, and horses may be carriers of Salmonella and may not shed the organism in their feces until stressed.35 Stress is difficult to define, and what may predispose one animal to disease if infected may not predispose the next. Various factors have been associated with increased prevalence of salmonellosis or the shedding of the organism in feces by equine populations, including transportation or shipping,14,56,81,82 surgery,14,83 feed withdrawal,14 change in feed,55 antimicrobial treatment,57,84–86 deworming,87 colic,57 and diarrhea.56 Some of these factors may be an outcome of the infection rather than predisposing factors, such as the association with diarrhea.
One of the first reports of salmonellosis in hospitalized horses was in 1969.83 Since that time, most outbreaks of equine salmonellosis occurred at veterinary teaching hospitals.32,33,54,88–91 The development of large equine veterinary hospitals in the twentieth century brought major advances in the health care available for equids, but also the congregation of horses with different levels of susceptibility to infectious disease agents and the likelihood for exposure to these agents.74 Hospitalized horses may be subjected to one or more of the factors associated with salmonellosis, including transportation to the hospital, altered diet, feed withdrawal before anesthesia, surgery, and treatment with antimicrobials.92 In addition to these stresses, hospitalized horses are often from multiple sources, and thus without existing biocontainment protocols, pathogens could be shared between animals from different sources. The occurrence of salmonellosis was a major driving force in the recognition of the need for infectious disease control programs in equine hospitals.74
Normal intestinal flora and motility likely make horses more resistant to colonization with Salmonella spp.93 Challenge studies that vary the doses of Salmonella organisms in horses subjected to feed change or restriction or to different antimicrobial regimens that may alter GI flora have not been reported. Fasting and diet change could alter the normal bacterial flora. Both stress and antimicrobial treatment may alter the normal flora in horses and thus predispose to colonization and eventually to disease associated with Salmonella infection.94 The barrier effect of the indigenous intestinal flora that prevents establishment and multiplication of potentially pathogenic bacteria is called colonization resistance.95 Although little is known about the complexity of interactions among indigenous microflora of the intestine in the horse, disturbances of the normal flora may predispose to colonization and multiplication of pathogenic bacteria.96 A syndrome of antibiotic-associated diarrhea is often associated with proliferation of different enteric pathogens, such as Salmonella spp., Clostridium difficile, and Clostridium perfringens.97
In one of the few experimental studies of the impact of various stressors on experimentally infected equids, Owen et al.14 reported that transportation of experimentally infected ponies resulted in reactivation of the Salmonella infection. Details regarding the duration and method of transportation are lacking in this report, and during transport the ponies also had feed deprivation as an additional stressor. The authors noted that treatment with oxytetracycline resulted in prolonged fecal shedding of Salmonella spp., in treated ponies. In another study of risk factors associated with isolation of Salmonella Saintpaul from hospitalized horses, those horses receiving antimicrobials were at increased risk for shedding the organism.98
Factors associated with hospitalization of equine patients were associated with salmonellosis decades ago.99 Factors associated with increased likelihood for nosocomial infection with Salmonella among horses hospitalized in an intensive care facility included large colon impaction, number of days on bran mash, duration of treatment with potassium penicillin, number of days that other horses were shedding the organism in the hospital, and mean ambient environmental temperature of 80° F (26.6° C) or more compared with 60° F (15.5° C).58 Factors associated with a reduced risk of infection included withholding feed. An increase in ambient temperature may increase risk of infection by favoring survival and multiplication of the organism in the environment, but also by compromising the immune response of the host.100 Most outbreaks of equine salmonellosis reported in the literature have occurred in summer months, reinforcing the concept that heat stress may make horses more susceptible to infection. However, the roles of environmental contamination and host resistance have not been evaluated separately. House et al.58 proposed that withholding of feed might reduce the risk of oral exposure to the organism by eliminating the possibility of consuming contaminated feed or feeding from contaminated surfaces.
PATHOGENESIS
Salmonella bacteria are transmitted by the fecal-oral route. In experimental studies, most animals require oral administration of very high numbers (≥108) of Salmonella organisms to cause disease.13 Before establishing colonization of the ileum and colon, ingested Salmonella cells must survive a series of host-derived obstacles, such as salivary bactericidal enzymes, stomach acid, intestinal proteases, lysozymes, antimicrobial peptides and bile salts, complement, and phagocytes, as well as interference by the normal bacterial flora, including nutrient competition and bacteriocins.7 Anything that interferes with the activity of these nonspecific responses is likely to decrease greatly the infectious dose of Salmonella required to cause disease, a process termed facilitation. For example, oral antacid preparations or administration of drugs that decrease gastric acid secretion increases the risk of salmonellosis, presumably from increased passage of viable bacterial cells into the intestinal tract.14,15 Similarly, antimicrobial treatment that disrupts the normal intestinal or colonic flora increases the risk of salmonellosis, along with several other causes of infectious enteritis.14 Many of the stress factors that predispose to salmonellosis, including transportation, sudden feed changes, spoiled feedstuffs, and other illnesses, probably act at least in part by affecting the innate resistance of the horse through the mechanisms previously described, for example, by disrupting the natural GI bacterial flora, reducing GI motility, or reducing the stomach acid or other natural barriers to colonization.
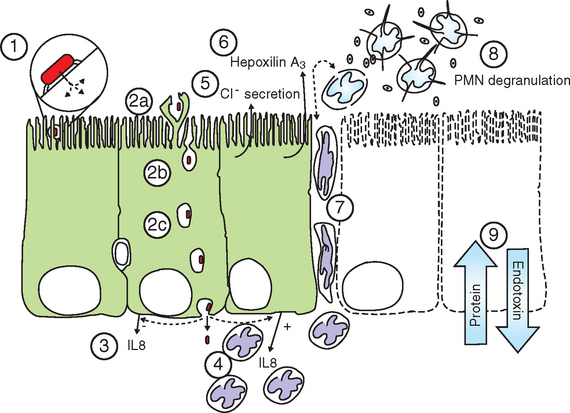
Invading cells that reach the target organs must penetrate the epithelial mucous layer and attach to the epithelial cell surface (Fig. 38-3). This attachment is mediated principally by bacterial fimbriae with specific receptor-ligand interactions with the epithelial cell surface.16 Genetic analysis and whole-genome sequencing of Salmonella Typhimurium revealed the presence of more than 10 different fimbrial systems.17 Most of these fimbriae are not expressed in culture but are activated in vivo. The multiple fimbrial systems probably work in concert, and the redundancy of fimbriae may be a strategy for Salmonella spp. to avoid immune responses directed at a single system.18
Salmonella spp. invade the intestinal epithelial cells by a process called bacterial-mediated endocytosis, which involves a rearrangement of the epithelial cell cytoskeleton triggered by proteins secreted by the attached bacterium, resulting in the epithelial cell membranes enveloping and internalizing the bacterial cell.19 However, invasion of epithelial cells may not be required for the pathogenesis of enteric salmonellosis. Most of the key events of salmonellosis, including neutrophil recruitment, intestinal inflammation, and increased fluid secretion into the intestinal lumen, are now known to be induced by Salmonella spp. through the actions of a “type 3 secretion system” (T3SS). Type 3 systems are molecular “syringes” that inject bacterial effector proteins directly into the host-cell cytoplasm and do not require internalization of the bacterial cell for their activity.20 The T3SS encoded in the “Salmonella pathogenicity island 1” (SPI1) plays a central role in bacterial-mediated endocytosis, and Salmonella isolates that lack T3SS-SPI1 are unable to induce the actin rearrangements that result in the internalization of Salmonella into the epithelial cells. In addition to invasion of epithelial cells, Salmonella cells also may traverse the epithelium by following phagocytes21 or may enter the submucosa directly through the disrupted epithelium characteristic of the villous tip where normal cell sloughing occurs.
During attachment and invasion, Salmonella T3SS-SPI1 effectors induce a massive recruitment of neutrophils. An important mediator of this process is the cytokine interleukin-8 (IL-8, now also termed CXCL8), secreted by epithelial cells in response to Salmonella infection. The specific trigger for IL-8 secretion is thought to result from an interaction of Salmonella flagellin with the toll-like receptor 5 at the epithelial basolateral cell surface.22 Flagellin may reach the basolateral cell surface along with invading Salmonella cells, but the proinflammatory effects of flagellin are also caused by its interaction with the T3SS-SPI1 effector SopE2.23 IL-8 and other chemoattractant molecules mediate the recruitment of neutrophils to the submucosal space. The subsequent diapedesis of neutrophils to penetrate the tight junctions between epithelial cells to reach the apical surface occurs in response to another chemoattractant whose secretion is triggered by T3SS-SPI1 effectors, the eicosanoid hepoxilin A3.24,25
Neutrophil chemotaxis to the apical epithelium, followed by degranulation and the release of inflammatory mediators, is largely responsible for the epithelial cell destruction and loss of epithelial barrier functions that elicit the clinical signs of salmonellosis.25–27 Release of inflammatory mediators by large numbers of activated neutrophils has long been considered central to the inflammation associated with salmonellosis.26,27 This inflammation results in massive epithelial damage, including sloughing of large areas of the epithelium with subsequent pseudomembrane formation. Inflammation and epithelial necrosis result in loss of serum protein into the lumen, with resultant hypoproteinemia typical of severe salmonellosis. Damage to the intestinal epithelium and the presence of invading salmonellae in the submucosa also result in release of endotoxin into the circulation, with the well-known subsequent effects on cardiac function, fever, leukopenia, hemoconcentration, lactic acidosis, coagulopathies, and hypotension (see Chapter 37).
In addition to their roles in epithelial invasion and neutrophil chemotaxis, T3SS-SPI1 effectors trigger increased fluid secretion at the site of infection. The increased fluid secretion is caused by increased chloride ion secretion, mediated in part by increases in the concentration of inositol polyphosphates (IPs) triggered by one or more T3SS-SPI1 effectors, likely including SopB. SopB contains motifs that are homologous to other IP-stimulating proteins and produces this effect on IPs in vitro.28,29
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree