Fig. 3.1
Replication cycle of influenza A Virus. 1 Attachment of influenza virus to sialic acid receptors on the cell surface. 2 Penetration of the virus into the cell through receptor mediated endocytosis. 3 Fusion of viral envelope with endosome membrane. 4 Uncoating and exit of the genome (vRNA) from virion into cytoplasm. 5 Entry of incoming vRNA into nucleus with the help of nuclear localization signals on certain proteins. 6 Synthesis of complementary RNA (cRNA) and replication of viral genome (vRNA). 7 Synthesis of mRNA from incoming vRNA. 8 Synthesis and glycosylation of envelope proteins (HA, NA). 9 Synthesis of other proteins. 10 Insertion of envelope proteins into the cell plasma membrane. 11 Assembly and budding of progeny virions. 12 Release of virions
Table 3.1
Summary of the roles of the proteins encoded by various segments in replication cycle of Influenza viruses
Genomic segments | Proteins encoded by various gene segments Influenza A virus | Role/Function in replication cycle influenza A viruses |
|---|---|---|
1 | PB2 | Binds the 5 ′ cap structure of cellular pre-mRNAs by the RdRp complex; Has nuclear localization signals for entry of RNPs into the nucleus for transcription and genome replication |
2 | PB1 | RNA polymerase function of RdRp complex; Has nuclear localization signals for entry of RNPs into the nucleus for transcription and genome replication |
3 | PA | Endonuclease activity of RdRp complex needed for host pre-mRNA cap-snatching; Has nuclear localization signals for entry of RNPs into the nucleus for transcription and genome replication |
4 | HA | Receptor-mediated endocytosis, HA2 component is the fusion peptide |
5 | NP | Has nuclear localization signals for entry of RNPs into the nucleus for transcription and genome replication |
6 | NA | Release of newly synthesized progeny virions from the plasma membrane |
7 | M1 | Nuclear export of newly formed vRNPs; assembly and budding |
M2 | Proton-selective ion channels and involved in endosomal fusion of endosome membrane with virus envelope; assembly and release | |
8 | NS1 | Interacts with various host factors |
NS2/NEP | Nuclear export of newly formed vRNPs; control of the accumulation of vRNA, cRNA and mRNA production; budding |
3.1 Viral Entry into the Host Cell
The recognition and binding of the HA with the host cell receptor molecule, terminal α-sialic acid on glycoproteins or glycolipids on the cell surface, is the first step of influenza virus entry (Sun and Whittaker 2013). The human and avian influenza viruses bind with sialic acid receptors with α-2, 6 linkage and α-2, 3 linkage, respectively. Both types of receptors are present in pigs (Skehel and Wiley 2000). This multivalent attachment by multiple copies of trimetric HA induces receptor-mediated endocytosis and subsequently the virus enters the host cell in an endosome. The viral envelope fuses with the endosomal membranes due the low pH of endosomes. This fusion causes acidic environment inside the endosome that leads to two events: conformational change in HA precursor polypeptide (HA0) and opening up the M2 ion channel. The buried HA2 fusion peptide is extruded to the distal tip of the HA spike as a result of pH induced conformational change in HA0. The newly formed HA2 N-terminal fusion peptide moves into the interior of HA trimer and connects with ionisable residues to produce a fusion competent neutral pH structure (Cross et al. 2009). The insertion of the fusion peptide into the host membrane induces juxtaposition of the two membranes. The merger of the two membranes occurs when the two ends of inserted HA2 come in close proximity, leading to the formation of a distinct hemifusion intermediate and the subsequent formation of a fusion pore. This fusion pore allows the release of the genomic segments of influenza virus into the cytoplasm of the host cell. Acidification of the viral core helps in the release of the viral RNPs from the endosome into the cytoplasm. The exposure of the virus to acidic pH within the lumen of the endosome, and subsequent protons flow into the viral interior, weakens the interaction of the M1 protein layer with the viral envelope and the RNPs. Prior to the fusion step, opening the proton-selective M2 ion channels allows protons to move through the viral envelope and acidify the viral core causing the virion to release the vRNP from M1 such that vRNP is free to enter the host cell’s cytoplasm (Pinto and Lamb 2006; Luo 2012).
3.2 Transcription and Replication of the Viral Genome
It is well known that Non-Coding Regions (NCR) have a key role in replication and transcription of influenza A virus. SMART (switching mechanism at 5 ′ end of RNA transcript) technology was used to identify the 5 ′ and 3 ′ NCRs of viral RNA (vRNA), complementary RNA (cRNA) and viral mRNA (Wang and Taubenberger 2013; Wang et al. 2014). There are some distinct differences in the transcription and replication of the influenza viral genome (Fig. 3.2). The transcription starts from a primer which is formed by 10–13 nucleotide long capped RNA fragments snatched from host pre-mRNAs. The premature termination of transcription occurs at a poly U domain which is copied by the polymerase to form poly A tail. The replication does not require a primer, and a full-length copy of the complete template is synthesized by viral polymerase as in this case it reads through the poly U region. Both the mRNAs and complementary RNAs (cRNAs) synthesised in influenza virus-infected cells are positive-sense RNAs. However, drastic differences have been observed in their structures. The mRNAs are capped, contain extra sequences of cellular origin at their 5 ′ -termini, lack viral sequences at their 3 ′ -termini and are polyadenylated. The cRNAs are neither capped nor polyadenylated and are complete copies of their vRNA (Benito and Ortín 2013). It has been shown that the transcription and replication of influenza virus is mediated by polymerases of different origins (Jorba et al. 2009).

Fig. 3.2
Differentiating features of transcription and replication of influenza viruses
The influenza virus RdRp, a heterotrimer composed of three subunits, PA, PB1 and PB2, is required for the synthesis of capped, polyadenylated mRNAs during transcription as well as full-length complementary RNA (cRNA) and genomic RNA (vRNA) in the cell nucleus during replication, using the vRNAs as template. Mature cellular messenger RNAs (mRNAs) have a 5 ′ methylated cap and a poly A tail. The vRNPs have poly A tails but no 5 ′ caps and the viral mRNAs have a 5 ′ methylated cap and a poly A tail, but the vRNA do not contain 5 ′ cap (Plotch et al. 1978). The transcription process of influenza virus is initiated by cap-snatching, i.e. the cleavage of 5 ′ -capped RNA fragments from host pre-mRNAs (Fig. 3.3). Subsequently, these are used as primers to copy the template and the mRNAs are finally polyadenylated. The promoter for mRNA synthesis by RdRp is situated in a partially double-stranded panhandle/corkscrew RNA structure formed by 13 and 12 conserved nucleotides at the 5 ′ and 3 ′ ends of the vRNA, respectively (Robertson 1979; Hsu et al. 1987; Flick et al. 1996; Brownlee and Sharps 2002). Previously it was thought that the cap snatching was done by PB1 or PB2 (Shi et al. 1995; Li et al. 2001; Guilligay et al. 2008). It has now been demonstrated that the endonuclease activity needed for cap-snatching to cleave cellular pre-mRNAs for the initiation of viral mRNA synthesis lies not on PB1 or PB2 but on PA subunit of the RdRp (Dias et al. 2009; Yuan et al. 2009). The PB1 subunit carries the actual RNA polymerase function. To make capped viral mRNA, the PB2 subunit binds the 5 ′ cap structure of cellular pre-mRNAs (Guilligay et al. 2008); N-terminal domain on the PA cleaves the cellular pre-mRNA ~10–13 nucleotide downstream of the 5 ′ cap producing a capped RNA primer (Dias et al. 2009; Yuan et al. 2009). This primer is then transferred to the polymerase active site on the PB1 subunit where it is used by PB1 as a primer to synthesize capped viral mRNA using the viral genes as templates.
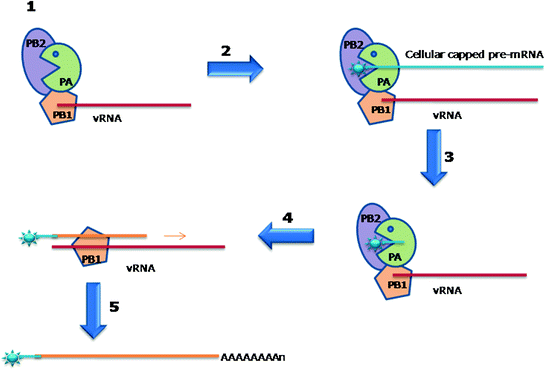

Fig. 3.3
Cap-snatching and transcription of influenza virus mRNA 1 Influenza virus polymerase complex binds with vRNA 2 Cap-binding domain located in the PB2 subunit of polymerase complex traps and binds with host pre-mRNA. 3 Endonuclease located in the PA subunit of polymerase complex cleaves approximaelty 10 nucleotides of host pre-mRNA. 4 The vRNA is used as a template to synthesise and elongate the chimeric vRNA by the nucleotidyl-transferase site in the PB1 subunit of polymerase complex. 5 Generation of a polyadenylated chimeric viral mRNA by polymerase stuttering mechanism
Viral mRNAs do not contain polyadenylation signal (AAUAAA) that is present in the cellular mRNA. Conserved sequence of five to seven U residues approximately 17 nucleotides from the 5 ′ end, are required for polyadenylation of virus-specific mRNAs (Robertson et al. 1981). As the bound RdRp reaches the 5 ′ terminus, its association with the 3 ′ NCR puts a steric blockage on the polymerase as it transcribes a uracil-rich region (Hay et al. 1977; Poon et al. 1998). This restricted mobility of the polymerase results in ‘stuttering’ mechanism leading to polyadenylation of the viral mRNAs by reiterative copying of the U(5–7) sequence due to back and forth movement of the RdRp over this stretch of U residues (Fig. 3.4). The end result of the entire process is the formation of a poly(A) tail (Poon et al. 1999).
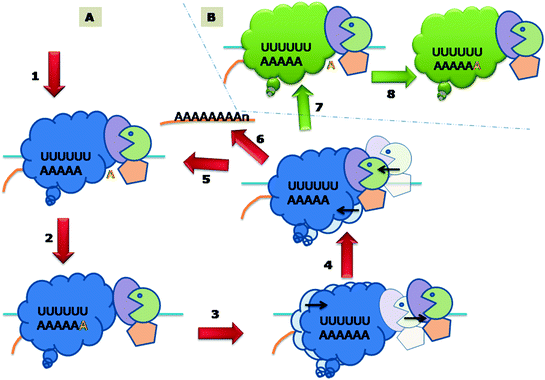

Fig. 3.4
Polyadenylation of influenza Virus mRNA by a Polymerase Stuttering Mechanism. A. 1 Binding of Transcriptase with Stop/Poly A signal on genomic template. 2 Insertion of the complementary nucleotide. 3 Forward movement of polymerase and growing mRNA. 4 Backward movement (Stuttering) of polymerase and growing mRNA. 5 Resumption of the steps 2, 3, and 4 again and again resulting in the addition of one A each time. 6 Or the newly synthesised polyadenylated mRNA is released. B. 7. 8. And the transcription of another genomic segment starts and the steps 2, 3, and 4 are repeated for this genomic segment
Various theories and mechanisms of replication of influenza viral genome have been reported. The viral RNA (vRNA) replication is completed in two stages. Initially the full-length complementary copy of the vRNA (cRNA) is formed, which is then copied to generate vRNA. Unlike the initiation of viral mRNA synthesis, the production of cRNAs and vRNAs does not require a primer. But the viral NP protein is required (Beaton and Krug 1986; Krug et al. 1989). The involvement and functions of NP in causing the shift between mRNA and cRNA synthesis has been studied from various viewpoints. Under the ‘encapsidation theory’ no regulatory function has been assigned to NP, except that it essentially acts as a co-factor to coat the nascent cRNA and vRNA segments (Shapiro and Krug 1988). The switch from capped RNA-primed transcription to primer-independent synthesis of cRNA and vRNA attributed to the RNA-binding activity of NP, purposed under the two models viz. ‘stabilization model’ (Vreede et al. 2004) and ‘template modification model’ (Portela and Digard 2002), have been discarded. This was based on the observation that efficient binding of the NP, which lack RNA-binding activity, directly to the viral polymerase can take place. It led to the proposal of another model, ‘polymerase modification model’ in which unprimed vRNA replication initiation was speculated to occur as a result of altered polymerase formed after the direct interaction between NP and the viral polymerase (Portela and Digard 2002; Newcomb et al. 2009). It has been reported previously that NP interacted with the basic subunits (PB1 and PB2), but not with the acidic subunit (PA) of polymerase complex (Biswas et al. 1998). The premature stoppage of RNA synthesis can be prevented by the NP binding to/or encapsidation of emerging/nascent RNA chains. Therefore, NP participates in the replication of viral genome both by RNA binding—and viral polymerase binding-dependent mechanisms. The host cellular nucleases will destroy the nascent cRNA unless it is stabilised by newly formed viral RNA polymerases and NP (Vreede et al. 2004).
Ubiquitin-specific proteases (USP) are involved in the deletion and processing of ubiquitin. A thorough proteomic analysis of purified influenza virus particles by complementary mass spectrometry revealed the presence of ubiquitin, one of the 36 host-encoded proteins, in influenza virions (Shaw et al. 2008). The RNAi libraries were put to high-throughput screening to detect cellular factors that may play some role in influenza A virus replication. The influenza A virus RNA replication was found to be regulated by deubiquitinase, USP11 but not by deubiquitinase USP10 or USP32. The USP11 is a host factor which interacts with the viral RNP complex through PB2, PA and NP proteins and has a role in influenza A virus RNA replication. The USP11 knockdown cells contained considerably elevated quantities of vRNA and cRNA as compared to the control cells. However, significant difference in the mRNA levels in the control and USP11 knockdown cells was not observed. The deubiquitinase activity of USP11 is responsible for the inhibition of influenza virus RNA replication, as its down-regulation resulted in increased virus yield, and viral genomic RNA replication was particularly prevented by USP11overexpression. The NP is a monoubiquitinated protein that can be deubiquitinated by USP11 in vivo by cleaving monoubiquitin from NP protein (Liao et al. 2010). The inhibition of the viral genome replication in K184 mutated NP indicated that the ubiquitination site of NP was K184 (Wasilenko et al. 2009) and revealed the critical importance of this residue for virus RNA replication (Liao et al. 2010). The RNA replication efficiency, but not transcription, can be increased by stabilization of cRNA due to modified interactions of monoubiquitinated NP with RNA. Further, the functions of RNP complex during RNA replication can be influenced by the interactions of cellular deubiquitinase USP11 with PB2, PA and NP.
The role of small viral RNAs (svRNAs) derived from influenza A virus in regulation of the switch from transcription to replication was also reported (Perez et al. 2010). The RdRp, NP and NS2/NEP are required for the generation of svRNAs, which are about 22−27 nucleotide long and match with the 5′ end of each of the vRNA segments. The synthesis of mRNA and complementary vRNA (cRNA) is barely affected by the depletion of svRNA, but a remarkable segment-specific deficit of vRNAs is observed. Further, expression of svRNA is associated with increased production and accumulation of vRNA. The transcription to replication switch is also influenced by the intracellular levels of nucleotides (Vreede et al. 2008).
Various host cell factors required for transcription and replication of the influenza virus genome has been described based on various studies on proteomic-based approaches (Mayer et al. 2007), interactomes networks (Bortz et al. 2011; Rodriguez et al. 2011; Tafforeau et al. 2011; de Chassey et al. 2013); knock down cells (Hsu et al. 2013); and RNAi screening (König et al. 2010; Karlas et al. 2010; Watanabe et al. 2010; Stertz and Shaw 2011; Bakre et al. 2013). The requirement for host factor(s) for the formation of vRNA has been demonstrated from the findings that the cRNA can be formed from incoming vRNA in infected cells, but not from the isolated vRNP. The viral RdRp enzyme produces predominantly abortive short RNA chains in the absence of influenza virus replication factor 1 (IREF-1)/minichromosome maintenance (MCM), and not the full-sized cRNA. The IREF-1/MCM helps in stabilizing the replicating polymerase complexes by supporting the interaction between the nascent cRNA and the PA subunit of the RdRp (Kawaguchi and Nagata 2007). Recently the replication of influenza virus was shown to be affected adversely by a newly identified host cellular protein HAX1 that acts by inhibiting the nuclear transport of PA. It was observed that the amount of PA was increased in HAX1-knockdown cells, and this PA accumulation in HAX1-knockdown cells could be reverted by re-expression of HAX1 protein (Hsu et al. 2013). The role of several human protein kinases (HPKs) and their regulation by miRNAs in the replication of influenza virus was recently reported (Bakre et al. 2013). The replication of the virus genome is followed by its encapsidation by NP and this encapsidation is facilitated by cellular protein, RAF-2p48/NPI-5/UAP56 (Kawaguchi et al. 2011).
3.3 Entry into and Exit of RNPs from Nucleus
Whether the released RNPs from the infecting influenza A virus are exported from cytosol into nucleus of the cells as one basket of eight RNPs or independently as separate RNPs is unclear. The essential nuclear localization signals (NLSs) for the entry of RNPs into the nucleus are present on all the proteins constituents (i.e. PA, PB1, PB2, NP) of the RNP complex. The residues located between 124–139 and 186–247 locations of PA (Nieto et al. 1994); between residues 187–211 of PB1 (Nath and Nayak 1990; Fodor and Smith 2004); and amino acids KRKR at 736–739 position of PB2 (Mukaigawa and Nayak 1991) were reported to possess NLSs. Two NLSs have been identified in the NP sequence. One of these was a typical bipartite NLS that was found between residue 198 and 216 with a sequence of 198RX13RKTR216 (Weber et al. 1998). A comparatively better accessible, non-conventional NLS (nNLS) with a consensus sequence of 3SQGTKRSYXXM13 at the amino-terminus of NP was also identified (O’Neill et al. 1995; Neumann et al. 1997; Wang et al. 1997; Wu et al. 2007a). However, the role of NP for RNP nuclear import is the most significant (O’Neill et al. 1995; Wang et al. 1997; Wu et al. 2007b). The NP mutated nNLS was unable to carry out the nuclear import of the RNP, which was also severely affected by competitive inhibition by short peptides that mimicked the nNLS (Cros et al. 2005).
The export of the newly formed vRNPs from the nucleus, for further virus assembly at the host cell membrane, is mediated by nuclear export signal (NES) carrying M1 and NEP proteins of influenza virus (O’Neill et al. 1998; Neumann et al. 2000; Bui et al. 2000). Neither NP nor any subunit of polymerase (PA, PB1, PB2) has any NES. The NLS located on the N-terminal portion of M1 is masked due to its binding to NEP. The M1 through its C-terminal region interacts with RNP as well as NEP (O’Neill et al. 1998; Baudin et al. 2001) and forms an RNP-M1-NEP complex. The vRNPs are exported out of the nucleus via the chromosome region maintenance 1 (CRM1) dependent pathway through the nuclear pores. The host CRM1 protein and viral activated cellular Raf/MEK/ERK (mitogen-activated protein kinase (MAPK)) signalling cascade are responsible for NES-containing protein/complexes exit from the nucleus (Fukuda et al. 1997; Elton et al. 2001; Pleschka et al. 2001; Boulo et al. 2007). The activation of the MAPK cascade occurs as a result of build-up of influenza A virus haemagglutinin in the host cell membrane before the assembled progeny virions at the cell membrane are released through budding (Marjuki et al. 2006). Recently, a second nuclear export signal (NES2) situated at N2 helix of the N-terminal region of NEP, was reported. The nuclear export functions of NES2 were similar to NES1 of NEP as it also required CRM1. The interaction between NEP and CRM1 is increased due to removal of the NES1 alone, while this interaction entirely stopped if the NES1 and NES2 parts are deleted. Both influenza A and B viruses have been shown to possess NES2 motif which is highly conserved (Huang et al. 2013). Additional functions of NEP have been detected and it has been implicated in the control of the accumulation of vRNA and cRNA as well as viral mRNA production. It is also involved in the efficient release of budding virions by bringing in a cellular ATPase to the cell membrane (Paterson and Fodor 2012). The vRNAs are exported individually and initially remain separated in the cytoplasm before assembling together in the cytoplasm in a microtubule independent manner. The Rab11 positive organelles, but not the viral proteins HA or M2, are essential for the co-localisation of different viral RNAs in the cytoplasm (Chou et al. 2013). The location of the NP on the apical side of infected nuclei indicates polarised export of the vRNPs (Elton et al. 2005; Loucaides et al. 2009).
3.4 Translation of mRNAs into Proteins
The newly synthesised mRNAs are transported back from the nucleus into the cell cytosol for translation into proteins. The transmembrane proteins (HA, NA and M2) production begins in the cytosol. Simultaneously, glycosylation and folding of these newly forming polypeptide chains occurs in the endoplasmic reticulum. These proteins are further modified and then carried through the Golgi apparatus and the trans-Golgi network to the cell’s plasma membrane (Doms et al. 1993).
3.5 RNP Packaging, Assembly and Budding of Influenza Viruses
Only limited reports on the random packaging model are documented for the random packaging of viral genomic segments into virions (Enami et al. 1991; Bancroft and Parslow 2002). It is now consensually believed that specific signals are present in the viral segments that direct which segments are to be packaged into the virions (Smith and Hay 1982). The sequence-specific packaging signals for influenza virus genome have been identified in the 5 ′ and 3 ′ non-coding and the adjacent coding region sequences on each of the eight genomic RNAs (Hutchinson et al. 2010).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


