Fig. 1
Schematic diagram of the biliary macrophage-RCT pathway. Cholesteryl esters (CE) stored in lipid droplets of macrophage foam cells are hydrolyzed by neutral cholesteryl ester hydrolases. Lipid-poor nascent preβ-migrating HDL particles acquire free cholesterol (FC) from macrophage foam cells via ABCA1, whereas the ABCG1 transporter facilitates cholesterol efflux from macrophages to α-HDL particles. FC is esterified to CE within nascent HDL particles by lecithin-cholesterol acyltransferase (LCAT), thereby generating mature α-HDL. The liver selectively takes up HDL-associated CE via scavenger receptor class B member 1 (SR-BI) and excretes HDL-derived cholesterol into the bile as FC or as bile acids (BA) after conversion. An alternative nonbiliary route for the transfer of cholesterol from the blood directly to the intestinal lumen is thought to contribute to fecal macrophage-derived cholesterol excretion (not shown)
The efflux of macrophage cholesterol to HDL occurs by passive diffusion, facilitated by scavenger receptor class B type I (SR-BI) or, more efficiently, by a unidirectional mechanism mediated by the ATP-binding cassette transporters ABCA1 and ABCG1 [4]. Functional analysis of the cholesterol efflux capacity of serum may provide useful data to assess the impact of particular clinical and experimental situations that induce qualitative or quantitative changes in the biological activity of key physiological cholesterol acceptors, such as HDL particles and apolipoprotein (apo) A-I, which is the main protein component of HDL. Recent data [5] have revealed that the HDL-mediated cholesterol efflux ability from macrophages, a metric of HDL function, may be clinically relevant since it displays a strong inverse association with both the carotid intima-media thickness and the presence of angiographically confirmed coronary disease, independently of the plasma HDL-cholesterol concentration.
The in vitro cholesterol efflux method allows the determination of the rate of cholesterol efflux from cultured cells to plasma acceptors (see Fig. 2 for a schematic diagram of the method steps). Radioactive labeling of cholesterol and its incorporation in cholesterol-donor cells provides an effective and sensitive method for detecting total cholesterol efflux from cultured human monocyte-derived macrophages or mouse macrophages. However, it should be noted that the efflux of labeled cholesterol from the radiolabeled cells does not necessarily represent the release of cholesterol mass but rather reflect the bidirectional exchange of cholesterol. Sankaranarayanan et al. [6] showed that net movement of cholesterol occurs when the cholesterol donor cells are cholesterol enriched, but not when they are cholesterol normal. The fractional release of label from cholesterol-enriched cells provides an estimate of the “efflux efficiency” of the studied acceptor. However, to quantify the mass movement of the cholesterol, changes in the cell cholesterol mass must be measured [6]. Rodent macrophages expressing preferentially specific cholesterol efflux pathways are usually used in the efflux method. However, the regulation of cholesterol efflux transporters appears to differ among cells from different species. This is the case of cAMP, which strongly stimulates lipid efflux in mouse macrophages but not in human macrophages [7]. Differentiation of human monocytes into macrophages requires incubation of the cells for several days with different polarizing factors. This may affect the expression of ABC transporters in the differentiated macrophages and, accordingly, the cholesterol efflux efficiency of the generated cells [8].
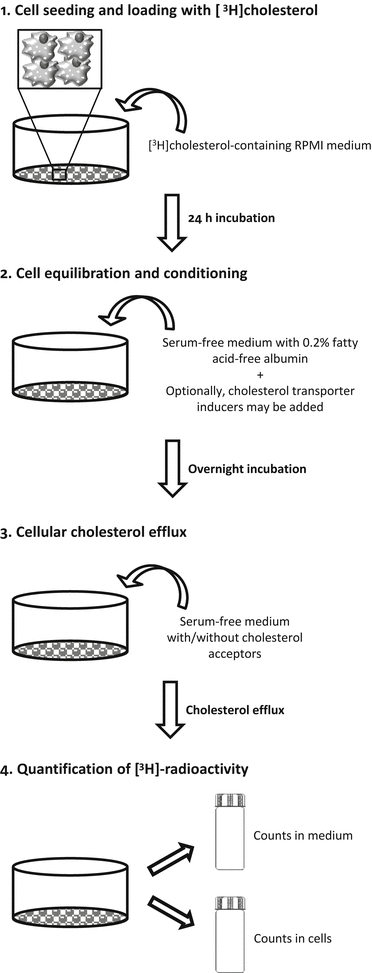

Fig. 2
Steps of cholesterol efflux method. 1: Macrophages are seeded and grown for two days in RPMI growth medium. If the starting cell typed are monocytes, the cells must be differentiated into macrophages by adding PMA to the macrophage growth medium. The differentiation of monocytes into macrophages typicallyFig. 2 (continued) stops the cells growing, and the cells are then prepared for radiolabeled cholesterol loading. Macrophages or monocyte-derived macrophages are incubated for 24 h with a loading medium containing radiolabeled cholesterol. 2: Excess radiolabeled cholesterol is then removed, and the cells are washed and incubated with a serum-free medium, which is supplemented with fatty acid-free BSA for 18 h to allow equilibration of the radiolabeled cholesterol with the intracellular cholesterol pools. If required, the cells can be treated during this step to modulate the expression of specific cholesterol transporters (cell conditioning). 3: After equilibration, the medium is removed, and the cell cultures washed before adding the cholesterol acceptors. The incubation of the macrophages in serum-free cell culture medium in the presence or absence of the acceptors for a specific period allows the estimation of the acceptor-induced release rate of radiolabeled cholesterol from macrophages. 4: The relative cholesterol efflux is calculated as the percentage of the [3H] counts released in the medium divided by the total [3H] counts (medium + cells)
For cholesterol efflux assays, cells are usually seeded in multiwell plates and maintained in growth medium until labeling with [3H]cholesterol. For cell labeling, [3H]cholesterol is added to the cells in growth medium, and the cells are incubated for 24 or 48 h, depending on the study. Cell labeling with [3H]cholesterol is followed by equilibration of the radiolabeled cholesterol among all intracellular cholesterol pools with serum-free medium, supplemented with fatty acid free-albumin for 18 h. When appropriate, the induction of the expression of specific cellular cholesterol transporters may be considered during this step. After equilibration, cholesterol efflux is measured by incubating the cholesterol-labeled cells in the presence of extracellular cholesterol acceptors. To calculate the relative release of acceptor-mediated radiolabeled cholesterol from the cells, the radioactivity in the medium corresponding to the effluxed cholesterol and that remaining in the cells is quantified. Although macrophages are most frequently used, the assay can also be applied to other types of cells, such as Fu5AH hepatoma cells and adipocytes, to obtain mechanistic information; Fu5AH hepatoma cells efflux cholesterol mainly via SR-BI and aqueous diffusion [9], and adipocytes efflux cholesterol via the SR-BI and ABCA1 but not the ABCG1 pathways [10].
The efflux of cellular cholesterol to HDL initiates RCT in all tissues, but the fraction that originates from the macrophage foam cells located in the intima is considered the only RCT component directly involved in atherosclerosis [2]. The key methodological problem for quantifying in vivo RCT from macrophages, which are the most important cholesterol-accumulating cell type in atherosclerosis, is that the macrophage cholesterol pool is a minor contributor to the total RCT from peripheral cells. To overcome this quantitative problem, Zhang et al. [11] developed an assay for measuring in vivo macrophage-to-feces RCT (macrophage-RCT) by tracing the reverse [3H]cholesterol transport from lipid-laden macrophages to feces in mice. In this assay, mouse macrophages or macrophage-like cells were loaded with acetylated low-density lipoproteins (LDL) and [3H]cholesterol and then injected intraperitoneally into recipient animals. After injection, the appearance of [3H]tracer in serum was determined at different time points and, most importantly, collected continuously in the feces during the time frame of the assay. The time course of such experiments is usually 48 h. This method distinguishes between the amount of fecal radioactivity in neutral sterols and bile acids (a schematic outline of this methodology is depicted in Fig. 3). The method can be used to study the role of different therapies and pathways relevant for RCT and HDL-mediated atheroprotection [12, 13]. Under certain experimental settings, the macrophage-RCT rate should be measured within a time spam shorter than 48 h. Therefore, a modified method has been used with pharmacological studies involving short-term transient activation of in vivo events that affect the cholesterol efflux capacity of HDL or acute pathophysiological conditions induced in mice [14, 15]. To evaluate the macrophage-RCT rate within a short time (i.e., in a period not sufficient to support the transfer of macrophage-derived cholesterol into feces), the tracer radioactivity in the intraluminal intestinal contents can be quantitated. The assay follows the same steps used in radiolabeling, injection of the macrophages, and processing of serum and liver samples. The only difference is that the macrophage-derived radioactivity of the intraluminal contents of the intestine is quantitated rather than that of the excreted feces. It should be noted that these macrophage-specific methods do not measure the net flux of cholesterol mass from the macrophages, but rather the unidirectional efflux of the radiolabeled cholesterol from the macrophages to plasma and the intestinal contents or feces. Moreover, this outward flow may be affected by exchange with endogenous tissue cholesterol pools. In addition, although the labeled macrophages in these assays are injected into the peritoneal cavity, which is considered a surrogate of the intimal space, the amount of the cholesterol acceptors in the intraperitoneal compartment and in the arterial intima may differ. The macrophage-RCT method described above has also been applied by injecting the labeled macrophages into extra-peritoneal compartments, such as the skin. However, the RCT fraction that originates from macrophages injected subcutaneously (e.g., in the rostral back), into the mouse requires longer time to be transferred to the intestine/feces in comparison with the faster macrophage-RCT rates obtained when the labeled cells are intraperitoneally injected [15]. Injecting labeled macrophages in other body locations, such as the tail skin, also appears to render a much less robust RCT signal [16].
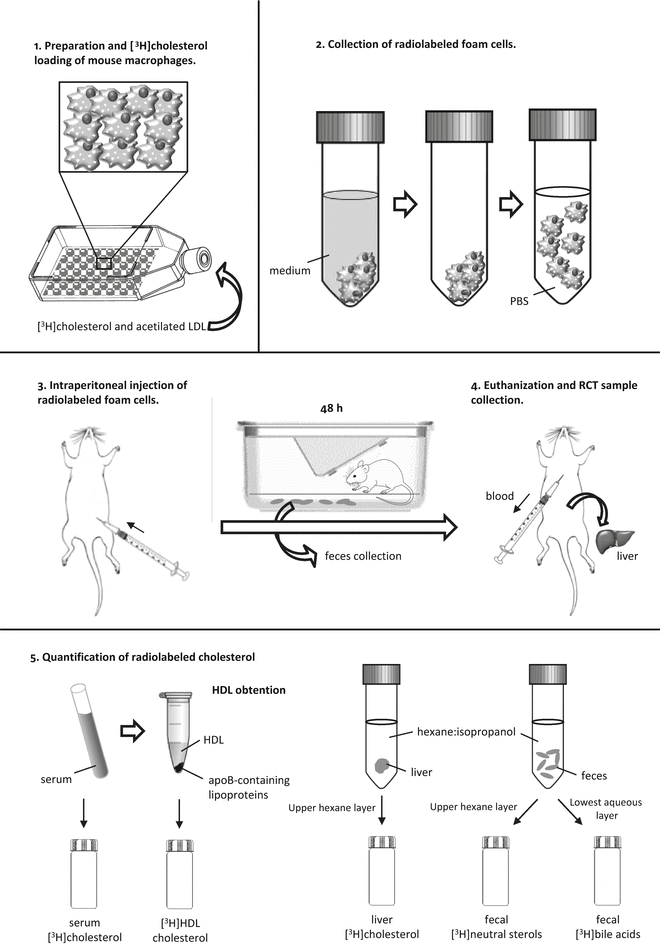

Fig. 3
Steps of the macrophage-to-feces RCT method. This method follows the fate of [3H]cholesterol from mouse macrophages injected into the peritoneal cavity of the mouse. 1: Mouse macrophages or macrophage-like cells are loaded with ac-LDL and [3H]cholesterol to become foam cells. 2: Radiolabeled foam cells are gently detached from the plastic surface of 75 cm2 flasks and resuspended in sterile PBS. 3: The radiolabeled mouse macrophages are then injected intraperitoneally into the mouse, and the mouse is placed inFig. 3 (continued) a cage with a metal grid floor for 48 h for feces collection. 4: At 48 h, the mouse is euthanized, the blood is collected by cardiac puncture, and the liver is removed. 5: [3H]-radioactivity is measured in serum and HDL after precipitating apoB-containing lipoproteins. Lipids from the liver and feces are extracted with hexane–isopropanol and partitioned against Na2SO4. Liver [3H]-radioactivity is determined in the upper layer, which contains the [3H]cholesterol. In the feces extract, the amount of [3H]-radioactivity is determined in the upper layer (neutral sterols) and the lowest layer (bile acids)
2 Materials
2.1 Cell Culture
1.
Human THP-1 monocyte cell line (ATCC® TIB-202™) and mouse J774A.1 (ATCC® TIB-67™) or P388D1 (ATCC® TIB-63™) macrophage-like cells.
2.
Roswell Park Memorial Institute medium (RPMI) 1640 supplemented with 2 mM glutamine.
3.
Growth medium for macrophages: RPMI 1640, glutamine, 10 % heat-inactivated FCS, 100 U/mL of penicillin, and 100 μg/mL of streptomycin (P/S) (see Note 1 ).
4.
75 cm2 cell culture flasks.
5.
Multiwell cell culture plates.
6.
10 mL serological pipettes.
7.
70 % ethanol in water.
8.
Sterile scissors and forceps.
9.
Dissecting board.
10.
Sterile phosphate-buffered saline (PBS).
11.
10 mL syringe and 25 G needle.
12.
50 and 15 mL polypropylene tubes.
13.
Cell scrapers.
14.
Sterile filters with a 0.2 μm pore size.
15.
Counting chamber (Fast read 102, Biosigma).
16.
Prepare Difco™ fluid thioglycollate medium (Becton Dickinson Labware) (3 % w/v) and autoclave for 15 min at 121 °C. Place the aliquots in 50 mL bottles. Keeping the solution in a dark room at room temperature (RT) for 1–2 months before use will increase the yield of inflammatory cells. Ensure that the broth is clear. Any cloudiness indicates contamination.
17.
1 mg/mL of phorbol 12-myristate 13-acetate (PMA) (Sigma), stored at −80 °C until use.
18.
10 mg/mL of 8-(parachlorophenylthio) cyclic-3′, 5′-AMP (CTP-cAMP; Sigma) in water, stored at −20 °C until use.
19.
Liver X receptor agonist (1 g/mL) T0901317 (Cayman Chemical) in ethanol, stored at −20 °C until use.
2.2 Macrophage Radiolabeling and Injection
1.
ApoA-I (Sigma) and HDL isolated by sequential ultracentrifugation at a density gradient of 1.063–1.21 g/mL from total plasma (or serum). Although the apoA-I prediluted with serum-free RPMI 1640 medium can be used directly, the isolated HDL must be desalted and incorporated into serum-free RPMI 1640 medium before using it. Incorporate the HDL into RPMI 1640 using a PD-10 desalting column, eluted by gravity. Before adding 2.5 mL of HDL, equilibrate the column with 25 mL of serum-free RPMI 1640 medium. Elute the mixture with 3.5 mL of RPMI 1640, and collect the whole eluate, which contains the HDL, in a separate tube. Whole plasma (or serum) or apoB-depleted plasma (or serum) can be used as acceptors to measure the efflux of the macrophage cholesterol.
2.
40–60 Ci/mmol [1α,2α(n)-3H]cholesterol (Perkin Elmer).
3.
0.2 N of NaOH.
4.
PD-10 desalting column (GE Healthcare).
5.
Ultracentrifugation tubes.
6.
Human LDL: isolate by sequential ultracentrifugation at a density of 1.019–1.063 g/mL and dialyze at 4 °C against 10 mM Tris–HCl, 1 mM EDTA, pH 7.4.
7.
Lipoprotein-depleted serum (LPDS): isolate by sequential ultracentrifugation at a density ≥1.21 g/mL and dialyze at 4 °C against 10 mM Tris–HCl, 1 mM EDTA, pH 7.4. To inactivate the LPDS, place it in a 56 °C water bath for 1 h.
8.
Acetylated LDL (ac-LDL): adjust the isolated LDL to 2 mg/mL of LDL protein (i.e., apoB or total protein content), and add an equivalent volume of saturated sodium acetate. Next, add 1 μL of acetate anhydride, and gently shake the mixture in an ice-water bath for 30 min. Add another 1 μL of acetate anhydride, and stir the mixture for 30 min. Finally, dialyze the mixture at 4 °C against 10 mM Tris–HCl, 1 mM EDTA, pH 7.4. Before using the mixture for the radiolabeling of mouse macrophages, incorporate the ac-LDL into RPMI 1640 using a PD-10 desalting column, eluted by gravity. To do so, equilibrate the PD-10 column first with 25 mL of RPMI 1640 before adding 2.5 mL of ac-LDL. Elute with 3.5 mL of serum-free RPMI 1640 medium, and collect by gravity the ac-LDL-containing eluate into a separate tube. Filter the suspension through a sterile filter with 0.2 μm diameter pores to avoid aggregated ac-LDL. Ensure that the ac-LDL suspension contains approximately 0.7–1 mg/mL of apoB.
9.
Absolute ethanol (>99.9 %).
10.
Bovine serum albumin (BSA), essentially fatty acid-free.
11.
2 mL plastic Pasteur pipettes.
12.
Liquid scintillation fluid (Optiphase Hisafe 2, Perkin Elmer).
13.
Scintillation 20 mL vials.
14.
0.4 % Trypan blue solution (w/v).
15.
1 mL syringes and a 26 G needles.
2.3 RCT Sample Collection and Quantification
1.
Isoflurane.
2.
Scalpel blade (Carbon steel 21, Swann-Morton).
3.
1.1 mL, Z-gel microtubes for serum separation (Sarstedt).
4.
Phosphotungstate precipitant reagent: 0.44 mM phosphotungstic acid and 20 mM MgCl2 (for precipitating apoB-containing lipoproteins).
5.
Reusable feeding needle—20 G/30 mm long.
6.
Solvent for lipid extraction: hexane–isopropanol (3:2, v:v). Store in a glass sealed bottle.
7.
Solution of 0.47 M Na2SO4 in bidistilled water.
8.
5 mL glass tubes.
9.
Thin layer chromatography (TLC) plates, size 6 × 20 cm (Whatman).
10.
TLC running solvent hexane–diethyl ether–ethyl acetate (50:50:1.5; v:v:v).
11.
Iodine crystals.
12.
Cholesterol reference standard: 10 mg/mL of cholesterol (Sigma) and 10 mg/mL of cholesterol stearate (Fluka) in chloroform, stored at −20 °C until use.
3 Methods
3.1 In Vitro Cholesterol Efflux Assays
Established cell lines of monocytes are currently used (e.g., THP-1 cells) for cholesterol efflux assays. Alternatively, monocyte-derived macrophages may be used (see Note 2 ). Primary human monocytes should be freshly collected with serum containing medium, Ficoll-Hypaque®, or Lympholyte® gradients from whole blood (see Note 2 ). The macrophages can be generated by supplementing the growth medium with PMA (i.e., THP-1 cells). In the case of primary human monocytes, they can be supplemented with granulocyte-macrophage colony-stimulating factor (GM-CSF) or macrophage colony-stimulating factor (M-CSF). Various established cell lines of mouse macrophages (e.g., J774 or P388D1 cells) are used to assess the rate of cholesterol efflux to specific extracellular acceptors. Apart from these cell sources, primary mouse macrophages can be isolated from the resident peritoneal macrophage pool either directly or after the injection of thioglycollate broth into the peritoneal cavity of mice.
3.1.1 Seeding and Differentiation of Human THP-1 Monocytes into Macrophages
1.
Culture THP-1 in growth medium in 75 cm2 cell culture flasks, and keep in a humidified cell incubator (5 % CO2) at 37 °C. Refeed every 2 days with fresh growth medium by adjusting the cell density between 0.8 and 1.2 million cells per milliliter.
2.
Sediment the cells at approximately 200 × g at RT for 10 min. Aspirate the supernatant and resuspend cells in macrophage growth medium.
3.
Count the THP-1 cells in a counting chamber.
4.
Seed the monocytes at a cell density of one million cells per well (in a final volume of 1 mL) in 12-well plates (see Note 3 ), and allow them to differentiate into macrophages in the presence of 0.05 μg/mL of PMA (see Note 4 ) in a humidified cell incubator (5 % CO2) at 37 °C for 4 days. After differentiation, the cells can be directly used for cholesterol efflux assay (continued in Subheading 3.1.4; see Note 5 ).
3.1.2 Seeding of J774 or P388D1 Macrophage-Like Cells
1.
Mouse J774 or P388D1 macrophage-like cells are typically seeded and expanded in 75 cm2 flasks in macrophage growth medium and kept in a humidified cell incubator (5 % CO2) at 37 °C until use.
2.
Change the macrophage growth medium every 2 days. To do so, pour the macrophage growth medium from the flasks, and wash the cells twice with 10 mL of warm PBS. Following this step, add fresh, warm macrophage growth medium and keep in a humidified cell incubator (5 % CO2) at 37 °C.
4.
At a cell culture confluence of 90 %, the cells are ready for subculture. Wash the cells twice with warm PBS, and add the macrophage growth medium as described in Subheading 2.
5.
Gently scrape the cells from the plastic surface with a cell scraper.
6.
Place the cell suspension into a polypropylene tube. Count the cells in the counting chamber, seed at a cell density of 300,000 cells per milliliter (see Note 7 ) and add growth medium up to 1 mL per well in 12-well plates.
7.
Keep in a humidified cell incubator (5 % CO2) at 37 °C for 1 day (continued in Subheading 3.1.4).
3.1.3 Isolation and Seeding of Primary Peritoneal Macrophages
Resident peritoneal mouse macrophages can be isolated via an intraperitoneal injection of thioglycollate into the mice. This produces an inflammatory response, resulting in the release of large numbers of macrophages, which can then be purified. The peritoneal inflammatory cells can be recovered from euthanized mice at various time points after the injection. Davies and Gordon [17] previously described this process in detail. It should be noted that inflammatory macrophages may respond differently to several stimuli than the resident primary macrophages.
1.
Inject 2.5 mL of complete thioglycollate medium using a 26 G needle into the peritoneal cavity of the mouse.
2.
At 72 h, euthanize the mice by overdose of isoflurane. Pin the mice onto a dissection board, with their abdomens up. Clean the surface of the abdomen with 70 % ethanol.
3.




Perform a small off-center skin incision over the caudal half of the abdomen with scissors, and expose the underlying abdominal wall by retraction.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


