Fig. 1
Morphology of normal and apoptotic smooth muscle cell nuclei. (a) Control mouse carotid artery demonstrating normal nuclear morphology. (b) Carotid artery showing nuclear condensation (filled arrows) compared to normal nuclei (open arrows) on H and E staining. Scale bars represent 50 μm. From ref. [1] with permission
The most frequently used markers for apoptosis are terminal UTP nick end labeling (TUNEL) which assays unprotected DNA ends (Fig. 2), and antibodies to neo-epitopes that appear due to caspase cleavage, including caspases themselves (Figs. 3 and 4). However, the markers of apoptosis (for example TUNEL and cleaved caspase 3) measure frequency, mark different points or durations of the process, and may even be positive in cells long after the process has finished, for example if the apoptotic body is not cleared [5]. As in most cases we do not know how long apoptosis takes in vivo, and how much of the process is identified by the markers, we cannot ascribe rates of apoptosis. If either the duration of apoptosis or the length of time marked differs between tissues or even the same tissue under different conditions, then the measured ‘frequency’ may not be an accurate assessment of how much apoptosis is occurring.
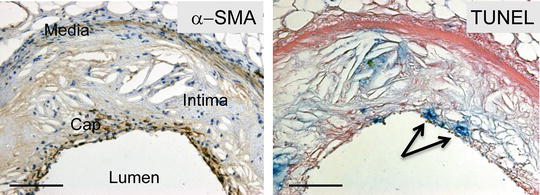
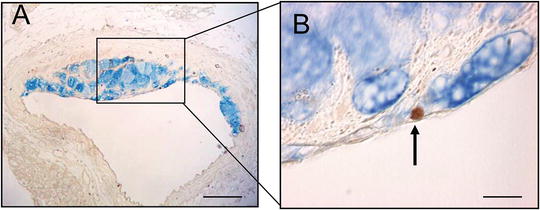
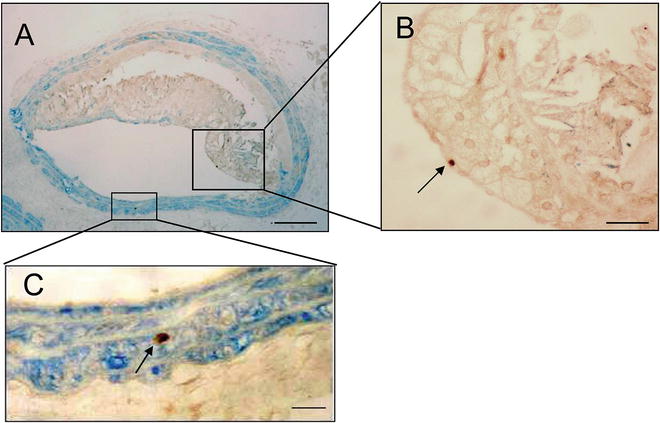

Fig. 2
Apoptosis of smooth muscle cells by TUNEL. α-SMA (left panel) and TUNEL (right panel) in mouse plaque. Arrows indicate TUNEL-positive cells in the fibrous cap. Scale bars represent 200 μm. From ref. [3], with permission

Fig. 3
Apoptosis of macrophages by cleaved caspase 3. (a, b) Double labeling for mac-3 (blue) and cleaved caspase 3 (brown) demonstrating apoptotic macrophage. From ref. [4], with permission

Fig. 4
Apoptosis of smooth muscle cells by cleaved caspase 3. (a) Brachiocephalic artery plaque double-labeled for cleaved caspase 3 (brown) and SMA (blue). (b) and (c), High power of area outlined in (a) showing caspase 3-positive, SMA-negative cell (b) or caspase 3 positive, SMA-positive cell (c) (arrows). Reproduced from ref. [4], with permission
This caveat is also important when ascribing changes in death frequencies to changes in apoptotic body clearance; clearance is a kinetic process that needs to be measured at least at two separate time points for a rate to be calculated, and assumes no change in generation of bodies, and no change in the duration of the process that is marked. We recently described a system whereby the kinetics of vascular smooth muscle cell clearance can be estimated in vivo. The system relies upon a single, defined, time-limited stimulus to induce apoptosis. We found that the half-life of apoptotic body clearance in the vessel wall was approximately 3 days, which could be delayed by challenging animals with a high-fat diet [6], confirming that the frequency of apoptosis observed depends upon the generation rate and the clearance rate.
Detection and particularly quantification of apoptosis generally requires a combination of techniques, as each is prone to artifact (see Notes 1 and 2 ), particularly in certain regions of the plaque where non-apoptotic debris collects, such as the necrotic core. Typical nuclear morphology together with TUNEL and/or cleaved caspases is usually sufficient. Identification of the lineage of the cell undergoing apoptosis is also difficult, as morphological features and lineage markers may be lost during cell death, and identification of a dead cell on adjacent sections is challenging (see Notes 3 and 4 ). Double labeling of apoptosis and a lineage marker is therefore preferred. Although apoptosis can also be detected in tissue sections using vital dyes (e.g., propidium iodide administered shortly before death [7]), or internucleosomal DNA fragmentation, these assays are not suitable for banked tissue, and therefore will not be considered here.
2 Materials
1.
Human and mouse paraffin sections mounted on glass slides suitable for microscopic examination.
2.
Proteinase K.
3.
Digoxigenin-dUTP.
4.
Cleaved caspase 3 antibody (e.g., Cell Signaling, cat. no. 9661).
5.
TUNEL assay kit (e.g., Roche Diagnostics, cat. no. 03 333 574 001). Contains:
(a)
Terminal Transferase (TdT) in 60 mM K-phosphate, 150 mM KCl, 1 mM 2-mercaptoethanol, 0.5 % Triton X-100, 50 % glycerol.
(b)
TdT Reaction Buffer 5× concentration: 1 M potassium cacodylate, 125 mM Tris–HCl, 1.25 mg/ml bovine serum albumin (BSA).
(c)
25 mM CoCl2 solution.
6.
Tris-buffered saline (TBS) buffer: 50 mM Tris–HCl, 150 mM sodium chloride, pH 7.6 (6.05 g Tris and 8.76 g sodium chloride in 800 ml of double distilled water, adjust pH, then make up volume to 1 L).
7.
Antigen unmasking solution, citrate-based (e.g., Vector Antigen Unmasking Solution, cat. no. H3300).
8.
Diaminobenzidine (DAB) solution.
9.
5-bromo-4-chloro-3-indolyl phosphate (BCIP)/Nitro blue tetrazolium (NBT) (e.g., Vector Kit SK5400). Prepared as follows:
(a)
To 5 ml of 100 mM Tris–HCl, pH 9.5 buffer add 2 drops of Reagent 1. Mix well.
(b)
Add 2 drops of Reagent 2. Mix well.
(c)
Add 2 drops of Reagent 3. Mix well.
10.
Phosphate-buffered saline (PBS) (e.g., Sigma, cat. no. D8537).
13.
Anti-digoxigenin alkaline phosphatase (anti-sheep F′ab fragments) (e.g., Roche Diagnostics: 11 093274 910).
14.
α-smooth muscle actin antibody (e.g., Dako M0851).
15.
Horse anti-mouse secondary antibody (e.g., Vector Laboratories BA2000).
16.
Alkaline phosphatase substrate III (e.g., Vector Kit SK-5300). Prepared as follows:
(a)
To 5 ml of 100 mM–200 mM Tris–HCl, pH 8.2–8.5 buffer.
(b)
Add two drops (80 μl) of Reagent 1.
(c)
Add two drops (80 μl) of Reagent 2.
(d)
Add two drops (45 μl) of Reagent 3.
(e)
Mix well before use.
17.




Aqueous mountant solution (e.g., VectaMount H-5000. Vector Laboratories).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


