Gene segments
Proteins encoded by various gene segments in
Influenza A virus
Influenza B virus
Influenza Ca virus
1
PB2
PB2
PB2
2
PB1, PB1-F2
PB1
PB1
3
PA
PA
P3
4
HA
HA
HEF
5
NP
NP
NP
6
NA
NA, NB
CM1, CM2
7
M1, M2
M1, BM2
NS1, NS2/NEP
8
NS1, NS2/NEP
NS1, NS2/NEP
The infection of the cells at high multiplicity of infection (M.O.I.) leads to the production of incomplete influenza virus particles (von Magnus virus) and the phenomenon is called von Magnus phenomenon after its discoverer von Magnus (1954). The genome of these incomplete influenza virus particles is defective, though their proteins and antigenicity is similar to the infectious viruses. The incomplete virus particles are either non-infectious or only partially infectious (Choppin and Pons 1970). These incomplete influenza virus particles are also called Defective Interfering (D.I.) particles as they are defective in their genome, require the help of complete influenza virus particles and interfere with the replication of the complete influenza virus particles (Nayak et al. 1978). The influenza D.I. particles possess small subgenomic RNAs (sgRNAs) derived by internal deletion of standard virus mainly of PB1, PB2, PA segments (Crumpton et al. 1978; Davis et al. 1980; Nakajima et al. 1979; Ueda et al. 1980; Nayak et al. 1982; Nayak 1983; Sivasubramanian and Nayak 1983). The M.O.I.-dependent production of influenza D.I. particles can occur due to defects in the maturation in which case all the newly synthesized set of genomic segments are not assimilated into infectious progeny virions (Lerner and Hodge 1969). The other possibility can be defective synthesis of various RNA segments of the virus within the infected cells (Nayak 1972). The formation of D.I. particles is not limited to influenza A viruses. The production of D.I. influenza B virus by a high multiplicity infection has been reported. These D.I. influenza B virus particles lack RNA segment 7 which codes for M protein and interfere with the replication of wild-type influenza B virus (Tobita et al. 1986).
The influenza A viruses are pleomorphic and can occur as spherical or filamentous forms. The diameter of the spherical virions is in the range of 80–120 nm. The virions isolated from fresh clinical isolates are predominantly filamentous with elongated viral structures >300 nm (Bourmakina and Garcia-Sastre 2003; Burleigh et al. 2005; Elleman and Barclay 2004; Wright et al. 2007). The morphology of influenza B virus is similar to that of influenza A virus. Influenza C viruses exhibit hexagonal reticular structures on the surface (Apostolov and Flewett 1969) and can form long (500 μm) cord-like structures on the surface of infected cells (Muraki et al. 2004; Nishimura et al. 1990, 1994). They are enveloped viruses having 10–12 nm long surface projections or spikes. There are two types of spikes in influenza A and B viruses, (i) rod shaped made up of homotrimers of haemagglutinin (HA) and (ii) mushroom shaped made up of homotetramers of neuraminidase (NA) glycoproteins (Cox et al. 2000). A typical schematic structure of influenza A virus is shown in Fig. 2.1. In contrast to influenza A and B viruses, influenza C viruses have only one spike that is made up of the multifunctional haemagglutinin-estrase-fusion (HEF) glycoprotein (Nakada et al. 1984, 1985). The envelope of influenza A virus has HA, NA and matrix 2 (M2) proteins whereas influenza B virus has HA, NA, NB and BM2 while influenza C virus has HEF and CM2 proteins (Betakova et al. 1996; Brassard et al. 1996; Cox et al. 2000; Nakada et al. 1984, 1985; Odagiri et al. 1999; Pekosz and Lamb 1997). Various viral and biological properties that differentiate the influenza A, B, and C viruses are shown in Fig. 2.2.
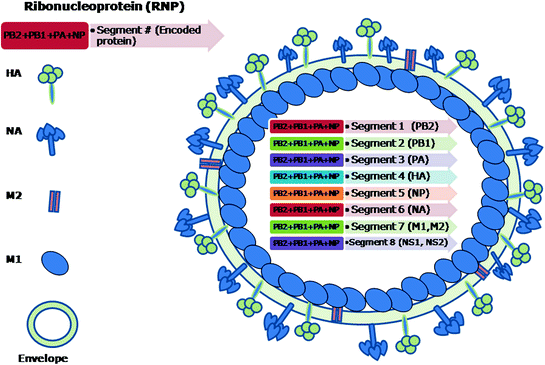
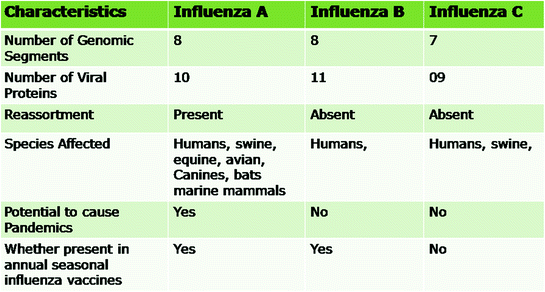

Fig. 2.1
Schematic structure of a typical influenza A virus. The genome of virus consists of eight segments of ssRNA which remain attached to polymerase complex proteins (PB2, PB1, PA) and wrapped in nucleoprotein (NP). The matrix protein (M1) is located underneath the lipid envelope. The M2 protein forms the ion channels. The haemmagglutinin (HA) and neuraminidase (NA) form the surface peplomers. The genomic segments 1–6 are monocistronic, while the 7th and the 8th are bicistronic. The NS1 and NS2 are considered to be non-structural proteins whereas the other six are structural proteins

Fig. 2.2
Differentiating features of various genera of influenza viruses
Influenza viruses are comparatively not very stable and do not survive for long in the environment, and physical factors such as heat, high or low pH and dryness can kill the virus. They are inactivated by organic solvents and detergents such as sodium desoxy cholate (SDC) and sodium dodecyl sulphate (SDS) (Swayne and Halvorson 2003). In the presence of organic matter, these viruses can be destroyed by chemicals such as formaldehyde, gluteraldehyde, beta propio-lactone and binary ethylenimine. After removal of organic matter, chemical disinfectants such as phenolics, quarternary ammonium compounds, 5.25 % sodium hypochlorite, 2 % sodium hydroxide, 4 % sodium carbonate dilute acids and hydroxylamine can destroy these viruses (Franklin and Wecker 1959; King 1991; Swayne and Halvorson 2003). Inactivation with the retention of haemagglutinating and neuraminidase activities can be achieved with various concentrations of formalin, binary ethylenimine and beta propiolactone for laboratory purposes (Laver 1963).
References
Alexander DJ (2001) Orthomyxoviridae—avian influenza. In: Jordan F, Pattison M, Alexander D, Faragher T (eds) Poultry diseases, 5th edn. W.B. Saunders, London, pp 272–279
Burleigh LM, Calder LJ, Skehel JJ et al (2005) Influenza A viruses with mutations in the M1 helix six domain display a wide variety of morphological phenotypes. J Virol 79:1262–1270PubMedCentralPubMedCrossRef
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


