Fig. 1
The programmatic structure of SACIDS visual centre
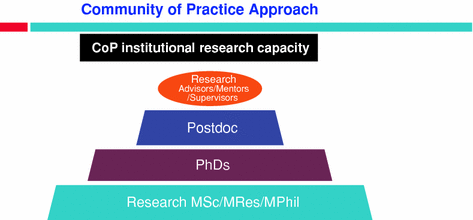
Fig. 2
RESEARCH APPRENTICESHIP through a community of practice approach

Fig. 3
Filovirus community of practice
3.2 The Role of NatCIDS
The National Centres for Disease Surveillance (NatCIDS) play a key national role in guiding, leading, training and generation of scientific innovations for use in the surveillance and risk management strategies including early warning systems, emergency preparedness and interventions needed for prompt containment or control of infectious diseases of humans and animals. NatCIDS are made up of relevant experts, epidemiologists and stakeholders across the country and have a principal role in promoting multi-institutional, multi-sectorial partnership and collaboration both at national and international level in order to implement common activities and achieve strategic objectives of SACIDS.
In brief, the main activities of NatCIDS could be summarised as follows:
To create a national platform to share and harmonise efforts from diverse sources with regard to infectious diseases surveillance and risk management
To assist country diseases surveillance programme by evaluating and strengthening surveillance systems both on medical and veterinary sides with specific reference to the “One Health Concept”
To work with stakeholders in preparing early, coordinated and accurate responses in case of epidemics or emerging disease occurrences
To provide a better understanding of the circumstances of infectious diseases occurrence through state-of-the art involvement and investigations during outbreaks
To source for resources through grants application for long self-sufficiency of the consortium at the national level
To create effective collaboration with other African countries and Northern partners in addressing One Health issues
3.3 Training for One Health
No country, rich or poor, is immune from the risk of microbial threats. Although specific conditions leading to emerging or re-emerging of infectious agents may be different, the challenge of managing disease spread is virtually the same, but not all countries have the same capacity to respond to this challenge. Within the “global village”, individual countries/regions have different priorities, often contrastingly varying financial and infrastructure resources. However, an unprecedented amount of rapid movement of humans, animals and animal products between these countries/regions presents a biorisk not only to an individual country/region, but to the entire international community. The growing realisation that pathogens do not respect traditional epistemological divides has resulted in the emergency of the ‘One Health’ initiative to advocate for closer collaboration across the health disciplines and has provided a new agenda for health education. Not only has the need for interdisciplinary participation been acknowledged in projects involving control of zoonoses (Marcotty et al. 2009; Roth et al. 2003; Zinsstag et al. 2007) but there has been a new awakening to change the way health professionals are educated (Marcotty et al. 2009; Zinsstag et al. 2011). It is against this background that SACIDS included in its capacity development programme the development of two One Health driven 2-year MSc programmes, one at the School of Veterinary Medicine, University of Zambia, specialising in analytical epidemiology and the second at Sokoine University of Agriculture in Tanzania, specialising in molecular biology (Fig 4).
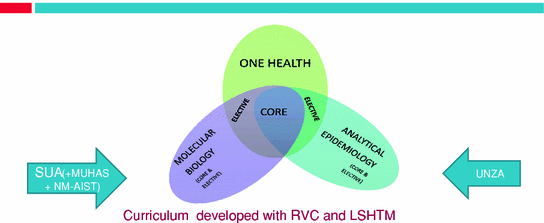

Fig. 4
Structure of SACIDS sponsored one health Msc At SUA and UNZA for Yr1 courses;(Yr2 devoted to research project)
3.4 Developing One Health Research Capacity
The Community of Practice (Wenger et al. 2002; Rweyemamu et al. 2010) approach for stimulating research within the SACIDS consortium is used to develop capacity in seven research themes, each with a specific exemplar. These have been (i) Emerging viral disease diseases—represented by Ebola and Marburg; (ii) Climate dependent, vector-borne diseases (Rift Valley fever); (iii) Diseases with potential for inter-species concern/spread between wildlife, livestock and humans—represented by Tuberculosis; (iv) Diseases of economic and food security importance—represented by foot-and-mouth disease; (v) Bacterial rare diseases—represented by plague; (vi) Systems for disease surveillance and preparedness analysis— focusing on participatory epidemiology and the application of mobile technologies to field data capture and transmission; and (vii) socio–economic approaches to One Health policy research.
The geographical focus of our studies has been in the ecosystems with a high human–livestock–wildlife interaction or in cross-border areas. The Community of Practice for each theme has comprised a career development postdoctoral fellow (supported for 3–5 years), two or three PhD students supported for 4 years and a similar number of M.Phil. or Research MSc students supported for 2 years, all co-supervised and mentored by the same pool of specialists from both Southern Africa and the UK or ILRI (Fig. 2). Each CoP has been encouraged to seek collaboration with other groups working in the same countries on related objectives.
4 Case Studies
4.1 Discovery and Characterisation of Novel Arenaviruses in Africa
Emerging diseases, notably zoonoses caused by negative-stranded RNA viruses continue to be a formidable problem for public health and veterinary communities globally. Arenaviruses, principally rodent-borne viruses, are capable of causing severe syndrome of viral haemorrhagic fevers (VHFs) in humans. The family Arenaviridae includes 23 recognised species, of which 6 can cause outbreaks of VHFs with high case fatality rates. Until recently, Lassa virus was the only known arenavirus to cause VHFs in humans in West Africa (Buchmeier et al. 2007; Charrel et al. 2008). However, the importation of a previously unrecognised arenavirus to South Africa after air medical transfer of a critically ill patient from Lusaka, Zambia in September 2008 resulted in a dramatic VHF nosocomial outbreak in Johannesburg with a case fatality rate of 80 %. International collaboration during this outbreak allowed for rapid identification of the novel virus, thus reassuring that health and scientific communities are committed and have powerful tools to rapidly detect and respond to the challenges of emerging unknown pathogens. The history of the outbreak, however, is a serious warning that highly pathogenic arenaviruses could be more widely prevalent in Africa (Paweska et al. 2009). It also evidences that international movement of patients contributes to unintentional spread of dangerous pathogens with dramatic public health consequences. Genome molecular study, including the application of unbiased high-throughput sequencing, allowed for rapid characterisation of a new member of the family Arenaviridae (Briese et al. 2009) provisionally named Lujo virus (LUJV), but our knowledge on its ecology, epidemiology, including host range, natural transmission cycle and distribution is in paucity. In the light of an unusual highly mortality rate in patients infected with LUJV, studies on tissue tropism, dynamics of viral replication and dissemination, mechanisms of pathogenesis in rodent host species and other potential vertebrate hosts, including primates, are needed.
The natural reservoir host of LUJV remains unknown likewise the source and route of infection of the first (index) case. She kept domestic pets and horses on her agricultural holding near Lusaka and there was evidence of rodent activity in the stables. Rodents could contaminate objects left unattended on the ground by excretion of virus in urine, including broken glass which apparently resulted in a deep cut to her foot about 10 days before medical evacuation to South Africa (Paweska et al. 2009). These circumstances might be of potential epidemiological relevance as she developed first symptoms within the incubation period of arenavirus infection after the cut. The natural reservoirs of arenavirus in Africa are rodents of the family Muridae, especially Mastomys natalensis. To-date only non-pathogenic arenaviruses have been found in areas surrounding Zambia. A study on the prevalence of arenaviruses among M. natalensis rodents in Zambia was conducted less than 1 year after the LUJV outbreak, from May to August 2009, including areas surrounding the cities of Lusaka, Namwala and Mfuwe, but not specifically on the farm of the index case. Nevertheless, of the total of 263 rodents captured, 5 were positive for an arenavirus infection in kidneys, of which 17 % of the 23 rodents captured near Lusaka and 4 % of the 24 captured in Namwala were positive, but none of the 143 rodents captured in Mfuwe were positive for arenavirus. Phylogenetic analysis of the four Zambian arenavirus isolates showed distinct sequences between Old World and New World arenaviruses. The novel Lusaka and Namwala strains, collectively designated Luna virus (LUNV), are genetically different from the LUJV, but closely related to nonpathogenic arenaviruses that have been found from central to eastern Africa (Ishii et al. 2011).
4.2 Re-emergence of Rift Valley Fever in South Africa 2008–2011
Rift Valley fever (RVF) was first reported in South Africa when a large outbreak occurred during 1950 and 1951. Subsequently, outbreaks with confirmed human cases occurred in South Africa in 1953, between 1974 and 1976, and in 1999. Limited and isolated outbreaks occurred in South Africa in 2008 and 2009 (Swanepoel and Paweska 2011; Bret et al. 2011), followed by a large, widespread outbreak during 2010 and 2011. Between 2008 and 2011, more than 2000 specimens from suspected human RVF cases were submitted to the Special Pathogens Unit of the National Institute for Communicable Diseases of the National Health Laboratory Service for laboratory confirmation. In 2008 and 2009, a total of 24 non-fatal human RVF cases were laboratory confirmed. In 2010, a total of 241 human cases were confirmed of which 25 were fatal. No human fatalities were recorded among 37 confirmed cases during 2011. The outbreaks were geographically linked with outbreaks in domestic ruminants and occurred mostly on the inland plateau of the country, notably in the Free State, Northern Cape, North-West, Eastern Cape and Western Cape provinces. The RVF outbreaks in humans peaked in March 2010 when more than 100 cases were laboratory confirmed. No new cases have been recorded in 2012 to date. In total, 302 human cases were confirmed from 2008 to 2011. In the same period, a total of 13,902 animal cases were confirmed of which 8,581 were fatal. Animal cases involved primarily domestic ruminants, but wild animals, including buffalo, sable, nyala, alpaca, llama and Asian buffalo were also affected. Overlap of geographic distribution of animal and human cases indicates that most humans were infected through direct contact with infected animals. Indeed, 254 (89 %) of the confirmed cases reported a history of contact with animal tissues or bodily fluids. All fatal cases occurred in 2010 (case fatality ratio 10 %). The overall case fatality ratio from 2008 to 2011 was 8 % (Jansen van Vuren et al. 2012). The 2008 and 2009 outbreaks were caused by the virus genetic variants representing lineage C comprising virus isolates from Zimbabwe (1978–1979), Madagascar (1991), Kenya (1997–1998, 2006–2007), South Africa (1999) and Madagascar (2003). The 2010 and 2011 outbreaks were caused by virus genetic variants related to a Namibian isolate from of lineage H first identified in 2004. One isolate from the 2010 outbreaks was genetically distinct, and was closely related to the Smithburn neurotropic vaccine strain. The virus was isolated from a veterinarian who experienced a needle stick injury while vaccinating livestock that likely was already infected by wild-type circulating virus. This isolate is therefore likely a reassortant of the wild-type and the vaccine RVFV strains (Grobbelaar et al. 2011).
4.3 Studying the Ecology of Filoviruses in the Congo Basin
The sporadic outbreaks of filovirus infections in humans are believed to result from contact with an infected animal and subsequent transmission between persons by direct contact with infected blood or body fluids. Infected individuals succumbing to filovirus infection exhibit virus-mediated impairment of early innate immune responses allowing for rapid progression of filovirus infection. The unavailability of antiviral therapy or approved vaccines, and the elusive nature of the spillover of filoviruses from a reservoir source to humans hamper countermeasures to effectively prevent the severe course of filovirus disease and transmission. Viruses belonging to the filovirus group were first discovered in 1967 for Marburg virus (Saijo et al. 2006) and in 1976 for Ebola virus (WHO 1978a, b). For a long time, the epidemiological circumstances surrounding filovirus outbreaks suggested that bats may have served as the primary source of infection in humans and non-human primates. Despite intensive efforts to trap thousands of vertebrate and invertebrate hosts in filovirus outbreak areas, isolation of live Ebola virus was unsuccessful, and the ecology and epidemiology of filoviruses are still not well understood. This is mostly because filovirus outbreaks occur irregularly, in poor resource, and remote areas of Africa, consequently investigations of filovirus outbreaks are hugely delayed and dependent on international support. In addition, handling of filoviruses requires the use of biosafety level four (BSL 4) facilities which are unavailable in these countries. Therefore, investigation of filovirus outbreaks, biology, ecology and epidemiology of filoviruses require a collaborative approach involving local researchers and overseas partners in order to secure required funding, training, diagnostic materials and laboratory support.
The SACIDS study of the ecology of filoviruses is undertaken in the Congo basin and particularly in the Democratic Republic of Congo (DRC) where several filovirus outbreaks occurred in the past. The programme is based on a collaborative approach involving several Congolese institutions (e.g. National Institute of Biomedical Research, Veterinary Laboratory of Kinshasa, University of Kinshasa), the National Institute of Communicable Diseases (NICD) a branch of the National Health Laboratory Services of South Africa plus the World Health Organisation (WHO). In this collaborative approach, the Congolese institutions provide administrative and human resources for sample collection in the field, the SACIDS, the NICD/NHLS and the WHO provide maximum security laboratory facility, training, financial and logistic support. Specific objectives of these studies are to collect samples from putative filovirus reservoirs in the DRC, notably from targeted bat species, and conduct their laboratory analysis in the BSL4 facility at NICD/NHLS (Fig. 2). These studies are based on the hypothesis of bats being reservoirs of Ebola virus and thus aim at isolating Ebola virus from bat organs.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


