Fig. 1.
Dispensing the PCR reaction mix into the port of the TaqMan miRNA fluidic card.
6.
Centrifuge twice at 1,200 rpm (331 ×g) for 1 min with maximum acceleration and deceleration (Fig. 2), seal the card (Fig. 3), and then trim the card (Fig. 4).
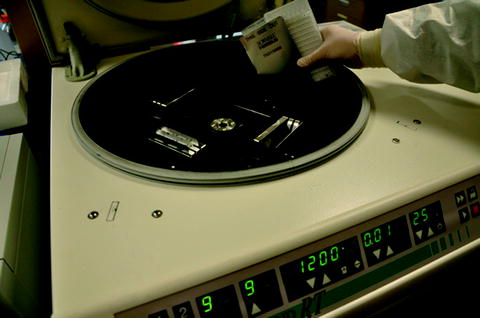



Fig. 2.
Loading the cards into the swing bucket adapters of the centrifuge.

Fig. 3.
Sealing the microfluidics card after centrifugation.

Fig. 4.
Trimming the card to fit into the PCR instrument.
7.
Import the appropriate SDS setup file (SDS.txt) located on the Information CD (supplied with the TaqMan miRNA fluidic card):
(a)
Start the SDS 2.3 software.
(b)
In the main menu, select File → New.
(c)
In the New Document dialog box, select the following from the drop-down menu:
Relative Quantification (ΔΔCt).
384-well TaqMan Low Density Array.
(d)
In the main menu, select File → Import to open the new document.
(e)
In the Open dialog box, navigate to the Setup.txt file specific for the array being run and click Import.
8.
Load the card into the 7900HT (Fig. 5) and run using the 384-well TaqMan Low Density Array default thermocycling conditions (10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min).


Fig. 5.
Loading the card into the 7900HT real-time PCR system.
3.7 Statistical Analysis
1.




RQ Manager is used to view results of the assay. The cycle threshold (CT) is the primary measure used to determine the original abundance of the specified miRNA in the sample. The CT is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. CT scores greater than 35 are considered nonspecific (undetectable) (8), so miRNAs in which 95% of individual observations had a raw CT score greater than 35 are excluded from the final data analysis.
< div class='tao-gold-member'>
Only gold members can continue reading. Log In or Register to continue
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


