Fig. 1
Atherosclerotic arteries harbor distinct immune cell infiltrates in plaques and adventitia. The adventitia of the normal arterial wall (left) constitutively contains vasa vasorum, lymph vessels, tissue macrophages, few T cells, mast cells, conventional dendritic cells (cDCs), and myofibroblasts. In response to transmural arterial wall inflammation during early stages of atherosclerosis, T cells and other immune cells such as B cells and monocyte-derived DCs (mDCs) infiltrate the adventitia. Throughout disease progression, extensive reorganization of adventitia segments adjacent to atherosclerotic plaques can be observed in the abdominal aorta of ApoE −/− mice resulting in ATLOs (right). Reorganization of the adventitia includes both the connective tissue-derived cells such as myofibroblasts and neogenesis of lymph vessels, angiogenesis, conduit formation, and high endothelial venule (HEV) formation. Concomitantly, monocytes are recruited through blood vessels, cDCs are recruited through lymph vessels, and naϊve T and B cells are recruited through newly formed HEVs. Moreover, survival niches are populated by plasma cells that secrete immunoglobulins in situ. In contrast to the adventitia, advanced atherosclerotic plaques are comparably acellular and show a limited set of adaptive immune cells. Adapted from Mohanta et al. 2014, Circulation Research, 114 (11):1772–1787
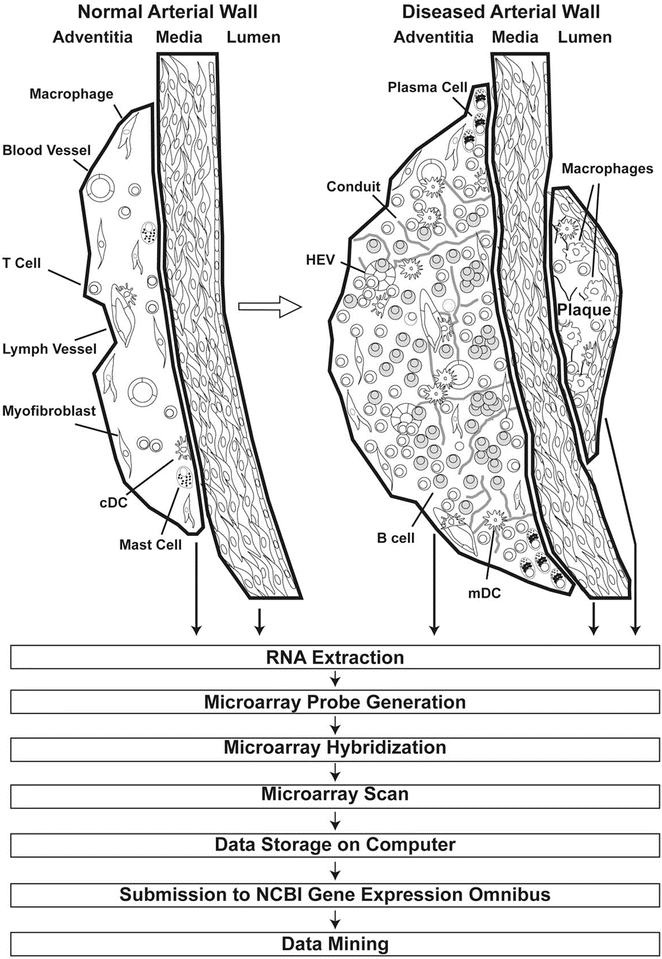
Fig. 2
Approach to construct transcript atlases of wild-type and ApoE−/− aortas from LCM-derived arterial wall laminae. Thick lines delineate the laser track to dissect and collect tissue compartments of the arterial wall laminae (see Fig. 1) from which total RNA is extracted for the preparation of mRNA expression microarrays. Subsequently, microarray probes are generated as described previously, hybridized to the microarray, scanned, and subsequently stored as CEL files in the NCBI GEO data bank. Various mining tools are available in the Internet to address a large number of issues; see [4–7]. Adapted from Mohanta et al. 2014, Circulation Research, 114 (11):1772–1787
2 Materials
2.1 LCM Procedure
1.
O.C.T.® compound (Sakura Finetek, Germany).
2.
Membrane slides (e.g., MembraneSlide 1.0 PEN, Carl Zeiss MicroImaging GmbH, Jena).
3.
Oil Red O (e.g., Sigma-Aldrich, Germany).
4.
Hematoxylin (e.g., Dako, Germany).
5.
Isopropanol.
6.
Acetone.
7.
PALM MicroBeam system (Carl Zeiss MicroImaging GmbH, Munich, Germany).
8.
Trizol (e.g., Invitrogen, Carlsbad, CA).
9.
Eppendorf cup (Eppendorf, Germany).
10.
Vacuum desiccator (e.g., Schott, Germany).
11.
Stereomicroscope (e.g., Carl Zeiss, Germany).
2.2 Preparation of RNA and Quality Control
1.
RNeasy® Micro Kit (Qiagen, Valencia, CA).
2.
NanoDrop ND spectrophotometer device (Thermo Fisher Scientific, Pittsburgh, PA).
3.
Agilent RNA 6000 Pico Kit (Agilent, Santa Clara, CA).
4.
Agilent 2100 Bioanalyzer (Agilent Technologies, Germany).
5.
Vortexer for Pico chip (e.g., IKA®-Werke GmbH & Co. KG, Germany).
6.
Ladder for RNA 6000 Pico chip (e.g., Ambion, Austin, TX).
7.
Chloroform.
8.
Ethanol.
9.
RNase-free H2O (e.g., DEPC H2O, Ambion, Austin, TX).
10.
Nonstick tubes (e.g., Ambion, Austin, TX).
2.3 Preparation of Microarrays from Small Amounts of RNA
1.
SuperScript II enzyme (Invitrogen, Carlsbad, CA).
2.
E.coli ligase (e.g., Invitrogen, Carlsbad, CA).
3.
RNase H (e.g., Invitrogen, Carlsbad, CA).
4.
T4 DNA polymerase (e.g., Invitrogen, Carlsbad, CA).
5.
T7-oligo(dT) Primer (e.g., Affymetrix, Santa Clara, CA).
6.
p(A) RNA (component of the RNeasy Micro Kit, Qiagen, Germany).
7.
ENZO™ High Yield In Vitro Transcription Kit (ENZO Life Sciences, Switzerland).
8.
Concentrated T7 polymerase (e.g., Ambion, Austin, TX).
9.
RNeasy® Mini kit (Qiagen, Germany).
10.
Phenol–chloroform (Ultra Pur) isoamyl alcohol (e.g., Ambion, Austin, TX).
11.
10× fragmentation buffer: 400 mM Tris-Ac, 1 M KOAc, 300 mM MgOAc.
12.
NH4Ac (e.g., Sigma-Aldrich, Germany).
13.
GeneChip® Hybridization Control Kit (Affymetrix, Santa Clara, CA).
14.
Control oligoB2 (Affymetrix, Santa Clara, CA).
15.
Herring sperm DNA (e.g., Promega, Madison, USA).
16.
Acetylated BSA, 50 mg/ml (e.g., Sigma-Aldrich, Germany).
17.
Tween-20 (10 % Surfact-Amps 20) (e.g., Thermo Fisher Scientific, Pittsburg, PA).
18.
GeneChip® Hybridization, Wash, and Stain Kit (Affymetrix, Santa Clara, CA).
19.
GeneChip® Mouse Genome 430A 2.0 Arrays (Affymetrix, Santa Clara, CA).
20.
GeneChip® Hybridization Oven (Affymetrix, Santa Clara, CA).
21.
GeneChip® Fluidics Station 450 (Affymetrix, Santa Clara, CA).
22.
GeneChip® Scanner (Affymetrix, Santa Clara, CA).
2.5 Mice
1.
ApoE−/− and wild-type mice (e.g., C57BL/6J background, The Jackson Laboratory, Bar Harbor, Maine, USA) (see Note 1 ).
3 Methods
3.1 LCM Procedure
1.
Euthanize mice.
2.
Perfuse the aorta in situ using EDTA/PBS for 10 min to remove blood taking care of preserving the adventitia.
3.
Remove adipose tissue and para-aortic ganglia but not LNs using forceps with the help of a dissection microscope (see Note 2 ).
4.
Embed aorta in Tissue-Tek and store at −80 °C.
5.
Heat membrane slides to 180 °C for 4 h to remove RNase.
6.
Store RNase-free membrane slides at −20 °C.
7.
Prepare fresh-frozen cryotome sections (10 μm) from aorta embedded in Tissue-Tek (maintained at −80 °C); use every tenth section for Oil Red O/hematoxylin staining to identify ATLOs or other arterial wall structures (see Note 3 ).
8.




Transfer five sections into Eppendorf cup to examine RNA integrity using Bioanalyzer electrophoresis (first RNA integrity control to test RNA quality before LCM) (see Note 4 ).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


