Fig. 3.1
The molecular clock in Drosophila: The schematic depicts the daily cycle that drives cyclic expression of per and tim mRNAs and PER and TIM proteins. Protein expression is tightly controlled by the indicated kinases and phosphatases. CRY is shown to reset the clock through its action on TIM. The interlocked loop that drives rhythmic expression of Clk mRNA is also shown. Note that while Clk mRNA cycles, the protein does not, so this loop may serve more to maintain stable levels of CLK rather than oscillations of clock components
After their translocation into the nucleus, around ZT18–22, the PER–TIM complex along with DBT binds CLK directly and represses transcription by promoting phosphorylation of CLK and releasing the CLK/CYC heterodimer from E-Boxes (Lee et al. 1998, 1999; Yu et al. 2006; Menet et al. 2010). The following morning, after lights-on, TIM is degraded (Hunter-Ensor et al. 1996; Lee et al. 1996; Myers et al. 1996). At this time, PER is again phosphorylated by DBT, but this time in the nucleus. Phosphorylated PER is recognized by SLIMB (an E3 ligase and targeted for ubiquitination-mediated proteosomal degradation) (Grima et al. 2002; Naidoo et al. 1999; Kloss et al. 2001). Subsequently, the newly formed hypophosphorylated CLK builds up and heterodimerizes with CYC to bind again to the E-box of PER to begin a new molecular cycle of the per and tim transcription.
In addition to this major feedback loop, most clocks contain a second transcription–translation loop in which one of the positive factors is regulated through negative feedback. In case of Drosophila, the positive factor regulated in this manner is CLK heterodimers bind to E-boxes present in the PAR domain protein (Pdp1e) and Vrille (Vri) and drive rhythmic expression of both these genes (Blau and Young 1999; Cyran et al. 2003) (Fig. 3.1). Vri and Pdp1e mRNA levels reach peak levels ~ ZT14, but the PDP1 protein product lags by about 6 h. vri mRNA and protein accumulate in phase with one another and VRI binds to the VRI/PDP1 box present in the Clk promoter region at ~ ZT14–ZT18 and represses Clk mRNA levels (Cyran et al. 2003). PDP1e levels rise several hours later and replace VRI on the V/P box to activate Clk transcription (Cyran et al. 2003; Zheng et al. 2009). This is facilitated by the action of PER–TIM. Around ZT18-4, the PER–TIM complex feeds back to inhibit CLK/CYC-dependent transcriptional activity, which results in reduced vri mRNA and protein expression and thus allows PDP1e to activate Clk transcription (Cyran et al. 2003). Around, ZT0–4 when the PER–TIM complex is degraded, a new cycle of vri and pdp1 transcription is initiated. VRI and PDP thus generate a loop to regulate a positive component of the major loop and thereby maintain ~24 h molecular oscillations (Fig. 3.1).
Mutants that abolish the transcriptional activity of CLK or CYC (Clk Jrk or Cyc 0 ) display peak levels of Clk mRNA, even though loss of CLK decreases expression of the Clk activator, PDP1, suggesting that there are additional activators of the Clk gene (Glossop et al. 1999). Genetic analyses of the Clk promoter region also support the idea that Clk can be transcriptionally regulated by transcription factors other than PDP1 and VRI (Gummadova et al. 2009).
3.3 Posttranscriptional Regulation
For most cycling components, rhythmic expression is brought about through transcriptional regulation. As noted above, a significant proportion of the genome is expressed cyclically, although the specific genes regulated in this manner may vary from tissue to tissue. However, in recent years it has become clear that circadian regulation can be exerted at several other levels also. These include intermediate steps from transcript to protein and can help cells adjust protein levels without altering the transcription process. Following mRNA synthesis, splicing and occasionally formation of multiple isoforms can occur. mRNAs are translocated into the cytoplasm where they undergo additional modification or degradation. Some of these steps include capping of the 5′ end, deadenylation of the 3′ end of the mRNA, splicing, and RNA editing. These molecular steps together can regulate stability of the mRNA and/or its translation.
3.4 mRNA Splicing and Stability
One of the most common posttranscriptional processes that regulates gene expression is alternative splicing, and, not surprisingly, this is involved in circadian regulation. For instance, alternative splicing in the eighth intron of the per gene allows for seasonal adaptation (Collins et al. 2004; Majercak et al. 2004). Shorter photoperiods as well as cold temperature induce alternative splicing events that affect PER expression such that per mRNA and protein accumulate earlier and to higher levels. On the behavioral level, the evening activity peak occurs earlier under cold temperature or short photoperiod conditions and the afternoon siesta is reduced (Majercak et al. 1999, 2004). Similar mechanisms have also been reported in Neurospora where per’s functional homolog, frequency (frq), also undergoes alternate splicing to adapt to seasonal variations, suggesting a conserved role of such molecular events (Colot et al. 2005; Diernfellner et al. 2005). The underlying mechanism is unknown, but it is speculated that trans-factors that regulate splicing are also controlled by seasonal variations. Thus, it appears that PER is the main determinant for integrating ambient day length and temperature information to the circadian clock.
A few earlier studies suggested additional posttranscriptional regulation of clock genes. For instance, the half-life of per mRNA in Drosophila varies throughout the circadian cycle (So and Rosbash 1997). Subsequent studies from mammalian labs showed that the mRNA stability of per2 and cry1 changes over time (Woo et al. 2009, 2010) and that regulated polyadenylation plays a critical role in circadian regulation. Using a genome-wide screen to identify mRNAs whose polyA tail exhibits circadian oscillations, Kojima et al. (2010) showed that the rhythmicity in polyA tails of several mRNAs positively correlates with rhythmic protein expression.
3.5 Role of miRNAs
Another potential route for posttranscriptional regulation is through miRNA-mediated RNA processing. miRNAs are about 22 nucleotides long/short noncoding RNAs that regulate expression of target genes by directly binding to the target transcripts and affecting RNA stability and/or translational efficiency (Bushati and Cohen 2007). miRNAs are generally characterized by their complementarity to 3′ untranslated regions (UTRs) of mRNA targets (Jackson and Standart 2007). A screen for cyclically expressed miRNAs in adult Drosophila heads from wild type and cyc 01 flies identified two miRNAs that are under circadian control—miR 263a and miR 263b (Yang et al. 2008). Predictions based upon specific algorithms indicated several core clock genes as potential targets, such as per, Clk, dbt, clockwork orange (cwo), and twins (tws) (Yang et al. 2008).
Using a different biochemical and genetic approach, Kadener et al. (2009) inhibited the miRNA biogenesis pathway in clock cells and followed up with tiling array analyses to identify novel miRNAs involved in the molecular clockwork. Three clock genes, Clk, vri, and cwo, were found to be associated with AGO1 (a catalytic unit of the RISC complex associated with miRNA biogenesis). Furthermore, reporter assays performed on the 3′UTR of the Clk gene identified bantam as an important miRNA, which is bound to this region as a part of the RISC complex (Kadener et al. 2009). Bantam was previously reported to have an essential role in stem cell maintenance in optic lobe development (Li and Padgett 2012). In vivo analyses of bantam also showed that it is required for proper maintenance of rest–activity rhythms.
Another miRNA implicated in circadian rhythms is miR-279, which was identified through a forward genetic screen (Luo and Sehgal 2012). Overexpression or knockdown of this miRNA impairs rest–activity rhythms without affecting the core clock; thus miR-279 functions as an output molecule. Specifically miR-279 regulates activity–rest rhythms through its direct effects on Unpaired (Upd), a ligand of the JAK/STAT pathway (Luo and Sehgal 2012). Together, these studies identify miRNAs mediated posttranscriptional regulation that has a key impact on the core clock or on the output in flies. Thus, miRNAs serve as major molecular players in circadian timekeeping, and future experiments warrant the identification and characterization of additional miRNAs in circadian regulation.
3.6 Translational Control
A recent mammalian study has shown that ~20 % of soluble proteins in the liver are regulated in a circadian manner, but their corresponding transcripts do not cycle (Reddy et al. 2006). This suggests an important role of posttranslational regulation in circadian rhythms. Such regulation has not been characterized in any detail in flies, but a few studies have indicated the possible relevance of translational control in the molecular oscillations of clock components. Translational control typically involves RNA-binding proteins, and one such protein, LARK, is implicated in the regulation of Drosophila circadian rhythms (McNeil et al. 1998; Newby and Jackson 1995; Huang et al. 2007). The lark mRNA cycles robustly and its expression is under control of the circadian clock (McNeil et al. 1998); however, the role of this gene is not fully understood. Through experiments to identify potential targets of LARK, Huang et al. (2007) identified Ecdysone-induced protein 74EF (E74). E74 mutants were shown to exhibit arrhythmic eclosion behavior, which corroborated earlier findings implicating lark in eclosion output. Mammalian LARK shuttles between the nucleus and the cytoplasm to control translation of target RNAs (Kojima et al. 2007; Lai et al. 2003; Lin and Fu 2007), and it is likely that Drosophila LARK does so as well. Interestingly many LARK targets contain miRNA binding sites (Ying et al. 2006), so it is possible that LARK acts through miRNAs to regulate the translation of specific target RNAs. A similar role has been shown for another RNA-binding protein, fragile-X mental retardation protein (FMRP), which has an important circadian function (Sofola et al. 2008; Dockendorff et al. 2002; Inoue et al. 2002).
Recent studies have shed light on the translational control of clock proteins. One particular study in flies showed that TWENTY-FOUR (TYF) is required for robust PER translation in the central clock neurons (Lim et al. 2011). TYF unlike LARK is not an RNA-binding protein, but is associated directly with polyA-binding protein (PABP) and indirectly with the 5′-cap-associating translation initiation factor eIF4G. Based on co-immunoprecipitation assays, per mRNA and, to somewhat lesser extent, tim mRNA was found with TYF. Interestingly in vivo, the loss of tyf affected only PER proteins but not the per mRNA. This study also showed that TYF was required mainly in the central clock neurons in the brain (Lim et al. 2011). Based on cell culture and in vivo data, it appears that following transcription, per mRNA is bound by a translational inhibitor, and TYF, together with PABP, eIF4G, and a few other factors, recognizes this inhibitor and displaces it. Taken together TYF ensures efficient translation of PER, although, as noted above, it is not yet linked to the delayed accumulation of PER relative to its mRNA.
3.7 Posttranslational Regulation
A transcriptional–translational loop would normally be complete in much less time than 24 h. For instance, subsequent to their synthesis, the PER and TIM proteins are translocated into the nucleus to repress their own transcription by inhibiting CLK/CYC transcriptional activity. These biochemical steps could be completed quite rapidly, and yet, they account for a significant proportion of a circadian cycle. Clearly, additional molecular steps introduce delays in the transcriptional–translational loop. The per and tim genes and their products not only cycle in their total abundance but also their phosphorylation state oscillates over the course of the day. This posttranslational modification of PER and TIM is very crucial for driving protein cycling and overt circadian rhythms, as the constitutive expression of per and tim mRNA can restore protein oscillations and behavioral rhythms in a significant proportion of per;tim double mutants (Yang and Sehgal 2001). In addition to phosphorylation, de-phosphorylation, formation of protein–protein complexes, and ubiquitination steps may also help to provide critical delays in feedback loops.
As mentioned above, CLK and CYC transcriptional factors heterodimerize and activate per and tim transcription during the day and into the early night. Any amount of TIM produced before sunset is degraded by light as TIM is sensitive to light; thus, TIM does not accumulate until dusk. Given that PER is unstable in the absence of TIM, due to its constant phosphorylation and degradation by DOUBLETIME (DBT) (Price et al. 1998), PER accumulation follows that of TIM. While DBT phosphorylates PER at several sites, phosphorylation at a specific serine residue (S47) is critical for targeting PER for proteosomal degradation (Chiu et al. 2008). S47-phosphorylated PER is recognized by the SLIMB protein, which is a component of the Skp1/Cullin/F-box protein (SCF) ubiquitin ligase complex, and targeted to the proteasome (Ko et al. 2002). Recent detailed studies have shown that PER phosphorylation involves several complex regulatory steps that affect either its stability or activity. For example, phosphorylation of the S589 residue is critical for PER stability and activity since it is involved in subsequent phosphorylation events, including phosphorylation at S47 (Kivimae et al. 2008). S589 phosphorylation inhibits DBT-mediated phosphorylation at S47 (Ko et al. 2002), thus producing more stable hypophosphorylated PER. However, this hypophosphorylated PER is less effective as a transcriptional repressor, and it is only upon de-phosphorylation of S589 that DBT-mediated phosphorylation generates a less stable but more transcriptionally activity form of PER (Kivimae et al. 2008). Furthermore, a recent study also shows that S589 does not act by itself in inhibiting phosphorylation at S47; rather it is part of a phospho-cluster which is important for modification of PER in a hierarchical manner. DBT action on this cluster is regulated by NEMO/NLK, a proline-directed kinase that phosphorylates PER at S596 to initiate a cascade of phosphorylation (Chiu et al. 2011).
Recent identification of two additional phospho-sites on PER, at 610 and 613, and their interaction with the per S domain further strengthens the idea that the per S domain acts as a major regulatory center (Garbe et al. 2013). Overall, these studies strongly indicate how the interplay of different kinases at phospho-clusters can generate a biochemical timer in Drosophila circadian clocks, since mutations at key phosphorylation sites lead to faster or slower clocks.
Posttranslational modification of TIM involves phosphorylation by GSK3β, which is called SGG in flies. Overexpression of SGG leads to period shortening accompanied by elevated nuclear TIM and PER levels, and reducing SGG levels results in period lengthening (Martinek et al. 2001). TIM can be directly phosphorylated by SGG under in vitro conditions and hypomorphic sgg mutants display reduced TIM phosphorylation (Martinek et al. 2001). Another important kinase, whose function in the molecular clock is somewhat unclear, is casein kinase 2 (CK2). CK2 is composed of two α and two β subunits, and independent mutations in these subunits result in circadian phenotypes in locomotor activity behavior (Akten et al. 2003; Smith et al. 2008). tik and andante are hypomorphic mutations of CK2 in the α and β subunits respectively that affect both PER and TIM stability and nuclear entry (Smith et al. 2008; Akten et al. 2003). These mutants exhibit long periodicity, reduced PER and TIM levels, delayed nuclear translocation, and longer circadian period under DD conditions (Smith et al. 2008; Akten et al. 2003). In vitro phosphorylation assays have shown CK2 can directly phosphorylate PER. On the other hand, reductions of CK2 activity in flies lead to elevated TIM levels even in the absence of PER, suggesting CK2 mainly targets TIM (Meissner et al. 2008). It is possible that CK2 acts as a priming kinase by phosphorylating PER and TIM and rendering them as imminent targets for DBT and SGG (Blau 2003).
The positive elements of the circadian clock are also posttranslationally regulated. Overall CLK levels do not cycle, but there are oscillations in CLK phosphorylation (Lee et al. 1998). CLK phosphorylation is coincident with the nuclear entry of the PER repression complex (PER–TIM–DBT) and so is implicated in the negative feedback mechanism (Yu et al. 2006, 2009; Kim and Edery 2006; Menet et al. 2010). It is thought that DBT (catalytically active or inactive) is required to recruit an unknown kinase that phosphorylates CLK (Yu et al. 2009). A recent study has shown that NEMO functions in clock cells to reduce CLK levels and to lengthen circadian period, but whether it directly phosphorylates CLK or not is not known (Yu et al. 2011). Another recent study reported that CK2 also targets CLK for phosphorylation leading to more stable but less transcriptionally active CLK (Szabo et al. 2013). Collectively these posttranslational steps are also interlocked across the different loops and provide key timing mechanisms in the molecular clock.
3.8 Anatomical Organization of Drosophila Circadian Clock
The molecular oscillations that give rise to circadian behavioral rhythms take place in distinct subsets of a total 150 neurons that are a small fraction of the ~100,000 neurons in the Drosophila brain (Fig. 3.2). These so-called clock neurons express canonical clock genes and are entrained by light. Some of the earliest studies by Handler and Konopka (1979) indicated that a small number of clustered neurons in the brain regulate adult locomotor activity rhythms (Fig. 3.2). The locations of these neurons were later determined by immunostaining of the per or tim gene products as well as their promoter-driven reporter activities (Zerr et al. 1990; Ewer et al. 1992; Kaneko et al. 1997; Kaneko and Hall 2000). However, per and tim mRNA and protein are also expressed outside the CNS, mainly in the compound eyes and ocelli, antennae, bristles, fat bodies, ovaries, prothoracic glands, and wings, where they generate peripheral clocks (reviewed in Helfrich-Förster 2002; Myers et al. 2003). per and tim genes are also expressed in glial cells, which lie in close proximity to clock neuron arborizations and are known to contribute to rhythmic behavior in a calcium-dependent manner (Helfrich-Förster 1995; Ng et al. 2011). However, they are not sufficient enough to maintain robust circadian rhythmicity in the absence of distinct clock neurons (Ewer et al. 1992).
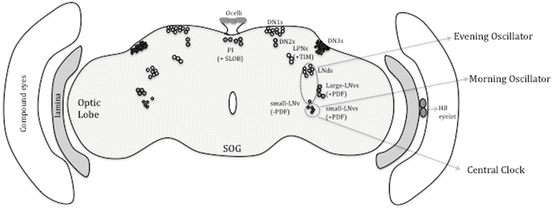

Fig. 3.2
The clock circuit in the brain. The different groups of PER or TIM expressing neurons in the brain are shown. Where functions have been well established, these are indicated. The neurons designated “central clock” are the small LNvs. These are necessary and sufficient to drive a rhythm in constant darkness
In adults, these neurons are bilaterally situated between the optic lobes and the central brain and broadly categorized into two subsets—lateral and dorsal neurons (Fig. 3.2; reviewed in Nitabach and Taghert 2008). The lateral neurons can be further divided into two subgroups on each side of the brain hemisphere. One group consisting of 6–8 is located dorsolaterally and is referred as LNd neurons. The other group consists of five small (s-LNvs) and four large (l-LNvs) neurons that are located ventrolaterally (Fig. 3.2) (Kaneko and Hall 2000). The small and large LNvs are defined by their small and large cell bodies. The dorsal neurons, as the name suggests, are located dorsally and they are composed of ~15 DN1, 2 DN2, and ~40 DN3 cells (Fig. 3.2). The DN1 and DN2s are medium-sized neurons and are located on the posterior side of the dorsal brain, while the DN3s are small sized that lie laterally in the dorsal brain region (Helfrich-Förster 2003). Besides these LN and DN clusters of neurons, there are additional neuronal clusters, called LPNs (lateroposterior neurons), that are PER and TIM positive (Fig. 3.2) (Kaneko and Hall 2000; Helfrich-Förster 2003). Several lines of evidence have shown that LNvs are the key pacemaker clock neurons, which are necessary and sufficient for maintaining circadian rhythms of rest–activity, in constant darkness (Frisch et al. 1994; Vosshall and Young 1995; Stoleru et al. 2004; Grima et al. 2004). On the other hand, the DNs have been shown to modulate and fine-tune the circadian rhythms under different environmental conditions (Murad et al. 2007; Stoleru et al. 2007).
The exact location and arborizations of the LNvs neurons (five small and four large in each brain hemisphere) could be defined through their expression of a distinct neuropeptide, pigment dispersing factor (PDF; Renn et al. 1999), with one exception that the fifth s-LNv does not express this peptide. Indeed, expression of the PDF peptide and the use of per- and tim-driven reporters established the composition of the clock network specifically the small and l-LNvs, LNds, DN1s, DN2s, and DN3s (Kaneko and Hall 2000). Except for the l-LNvs, the other clock neurons project towards the dorsal protocerebrum (Helfrich-Förster 2003). The l-LNvs connect to the accessory medulla via the posterior optic tract in the central brain (Fig. 3.3). Dorsal projections from the s-LNvs and projections to the optic lobes from the l-LNvs indicate that a common circadian output signal is relayed to different parts of the brain (Fig. 3.3) (Helfrich-Förster 2003). The dorsal area has more neuronal connections than other parts of the brain, suggesting that the circadian output signal may be transferred to different clusters of neurons in this region for timing neurophysiological functions (Fig. 3.3).
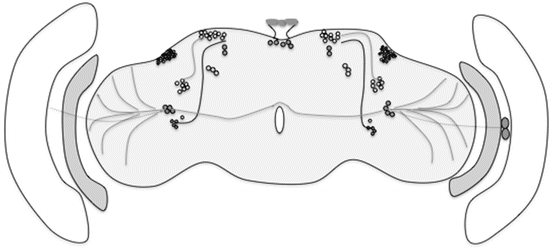

Fig. 3.3
Wiring of the clock circuit. The diagram depicts known projections of specific clock neurons. As might be evident, these have not been very well studied for the non-LNv clock cells. Within the LNv cluster, the l-LNvs project to the optic lobes and to the contralateral side and the s-LNvs project dorsally. In addition, the LNvs receive photic input from the Hofbauer–Buchner eyelet shown in the medulla
These 150 clock neurons expressing canonical clock genes are intricately networked and give rise to robust behavioral rhythms (Fig. 3.3). Earlier studies had indicated that LNvs might be the key pacemaker neurons for driving endogenous rhythms. Disconnected (Disco) mutants lacking LNs only but with intact DNs exhibit arrhythmic rest–activity as well as eclosion behavior (Dushay et al. 1989). In addition, genetic studies where PER function was rescued only in the LN clusters (all three subsets), but not in DNs, restored rhythmic behavior in per null flies (Frisch et al. 1994). Conversely, PER expression only in DNs did not result in robust rhythmic behavior (Ewer et al. 1992). Additionally, recent genetic analyses have definitively identified the s-LNvs as the main pacemaker neurons that drive circadian rhythms under constant dark (DD) conditions (Fig. 3.3), while l-LNvs remain largely dispensable under these conditions (Grima et al. 2004). For instance, rescuing PER function only in small LNvs but not in l-LNvs is sufficient to restore rhythmicity in flies lacking PER under DD conditions (Grima et al. 2004).
In LD conditions, the s-LNvs serve a special role in maintaining the morning peak of locomotor activity (Grima et al. 2004; Stoleru et al. 2004). The l-LNvs as described above have not been assigned any circadian function; however, recent reports strongly suggest that they are involved in detecting light input from visual and perhaps circadian photoreceptors (Sheeba et al. 2008; Shang et al. 2008). In brief these studies indicate that the l-LNvs are important for diurnal behavior, light-dependent arousal, as well as circadian phase advances (Shang et al. 2008). These subsets of neurons are ideally located in such a way that they receive light input signals from optic lobes and HB eyelets (extra photoreceptor important for entrainment) (Helfrich-Förster et al. 2002). Presumably, these neurons integrate rhodopsin-based light information along with information from the circadian photoreceptor, CRY (which is expressed in these neurons), to the circadian clock. Additionally the l-LNvs along with the DN2s apparently form a distinct circuit of unknown function (Stoleru et al. 2005) (Fig. 3.3).
The fifth s-LNv is similar in size to other s-LNvs but lacks PDF expression. This particular LNv and the LNds regulate the evening component of activity rest behavior under LD conditions (Grima et al. 2004; Stoleru et al. 2004). Thus, the PDF positive s-LNvs constitute the morning (M) oscillator and the fifth LNds + LNds make up the evening (E) oscillator. These studies also support the two-oscillator (M-E) model proposed by Pittendrigh and Daan (1976) which allows adaptation to the different photoperiods that occur at different times of the year. Briefly, the M oscillator exhibits a shorter period under LL conditions, whereas the E oscillator shows period lengthening and thus helps to adapt to the longer photoperiod by broadening the phase of activity. Then what role do DNs play? Although the s-LNvs are sufficient for maintenance of rhythms under DD conditions, the flies lack robust rest–activity behavior and show long period (Murad et al. 2007). The DN1s are presumably important for driving rhythms under constant light conditions (LL) (Murad et al. 2007; Stoleru et al. 2007). The DN2s lie in close proximity to the dorsal projections from the s-LNvs and are somewhat unusual in their molecular expression. The PER expression profile is out of phase when compared to the rest of the clock neurons, but whether this has any impact on circadian behavior is not yet known (Kaneko et al. 1997; Stoleru et al. 2005). The role of DN3s has also been studied. These neurons are not important for driving rhythms in DD, but under LD conditions, they are critical for the maintenance of the evening peak of rest–activity behavior (Veleri et al. 2003). Taken together, the overall role of the DN cluster is to provide robustness to the core pacemaker cells under different environmental conditions (Murad et al. 2007; Stoleru et al. 2007).
The lateroposterior neurons (LPNs) are another group of circadian neurons that are TIM positive but lack PER and do not quite belong to any other subset of clock neurons (Yoshii et al. 2005; Shafer et al. 2006). LPNs may function together with DN2s in temperature entrainment of activity–rest pattern (Miyasako et al. 2007). We can say that the regulation of circadian behavior emerges from a very complex network of clock neurons in which distinct group of cells have specialized roles in determining the rest–activity pattern under different environmental conditions. Furthermore, flexibility in the networking of clock neurons appears to facilitate adaptation to different environmental signals. In recent years, the power and advent of newer Drosophila genetic tools has added much valuable information to our understanding of the circadian regulation of rest–activity behavior.
3.9 Entrainment of Circadian Rhythms
How do circadian clocks respond to the outside world? To cope with the fluctuating external environment, circadian clocks have evolved sophisticated molecular pathways that help them to adapt to these conditions. Of the various environmental signals—light, temperature, humidity, social cues, and magnetic field—light is the most important zeitgeber (“time giver” in German) for entrainment of circadian rhythms. We will review a few key studies that elucidated the mechanisms by which circadian neurons synchronize to external signals.
Entrainment is achieved through classical ocular organs such as compound eyes, ocelli, or nonclassical pathways like the Hofbauer–Buchner (HB) eyelet and the blue-light photoreceptor CRYPTOCHROME (CRY) (Helfrich-Förster 2005). The compound eyes, ocelli, and HB eyelets use different sets of rhodopsins, whereas the canonical clock neurons express CRY. All of these photoreceptors contribute towards entrainment of the circadian clock. Flies missing one or two of these photoreceptors still retain the capability to synchronize to LD cycles, although they display impairments in specific circadian responses to light (Emery et al. 2000; Helfrich-Förster et al. 2001, 2002; Stanewsky et al. 1998; Klarsfeld et al. 2004). However, eliminating all photoreceptors including CRY renders the clock completely blind to the external environment, as it starts free-running even under entraining conditions (Helfrich-Förster et al. 2001; Klarsfeld et al. 2004).
The anatomical pathways that carry signals from the compound eyes and the ocelli to central clock neurons are not completely understood. The HB eyelets, which are located within the optic lobes, send axonal projections to the accessory medulla and have also been shown to make connections with LNvs (Malpel et al. 2004; Helfrich-Förster et al. 2002). The precursor of HB eyelets, Bolwig’s organs in larvae, is connected to the LNvs, and it gates the circadian rhythm of light avoidance, which requires the larval pacemaker neurons (Malpel et al. 2002; Helfrich-Förster 2002; Mazzoni et al. 2005).
CRY belongs to a family of flavoproteins and is expressed in most circadian neurons. Emery et al. (1998) found that cry b hypomorph mutants (a point mutation in the flavin-binding domain) render flies rhythmic in constant light (typically flies lose rhythms under these conditions) and significantly less sensitive to the light pulses (Emery et al. 1998; Stanewsky et al. 1998). Conversely, flies overexpressing CRY proteins showed increased behavioral responses to light pulses (Stanewsky et al. 1998; Emery et al. 1998, 2000; Egan et al. 1999). Expression of CRY solely in the LNvs in a cry b genetic background also substantially rescues the light responses, but completely restoration of the photic response is achieved through expression in a broader set of clock neurons (Emery et al. 2000). Expressing CRY just in the compound eyes does not rescue the circadian light response in cry b flies, suggesting that CRY acts in clock neurons. At the molecular level, CRY upon activation by light undergoes a conformational change and binds to TIM. This step, which leads to the degradation of TIM and subsequently CRY, is crucial in the daily resetting of the molecular clock at dawn (Suri et al. 1998). As a result PER becomes available for phosphorylation by DBT and is also degraded and a new cycle is initiated. We now know that degradation of TIM involves a specific E3 ligase, an F-box protein JETLAG (JET) (Lin et al. 2001; Koh et al. 2006), which promotes ubiquitination and subsequent proteosomal degradation of TIM (Naidoo et al. 1999; Koh et al. 2006; Peschel et al. 2006). SGG is another important protein whose overexpression mimics the behavioral phenotype of cry b in constant light (LL) conditions (Stoleru et al. 2007). Apparently, SGG binds with and stabilizes CRY and makes it unavailable for binding with TIM. This step results in loss of TIM degradation and persistent rhythmicity under LL conditions (Stoleru et al. 2007). As noted above, other phototransduction pathways can also entrain circadian rhythms. Double mutants cry b and norpA P41 , which lack CRY as well as a phospholipase C required for vision in ocular photoreceptors, can still entrain to LD cycles, indicating yet another mode of photic input into the clock (Helfrich-Förster et al. 2001). However, loss of function of the glass (gl 60j ) gene together with cry b renders flies completely blind (Helfrich-Förster et al. 2001). gl 60j flies lack all known photoreceptors including those in compound eyes, ocelli, as well as HB eyelets (Helfrich-Förster et al. 2001; Veleri et al. 2007). Furthermore, blocking synaptic transmission between the HB eyelet and its potential target cells, the LNvs, results in defects in behavioral resetting particularly under extreme photoperiods (Veleri et al. 2007). This study established the HB eyelet’s role as an additional photoreceptor in the circadian clock.
Flies have also evolved molecular mechanisms to fine-tune their circadian response to light. For example, a natural polymorphism in the tim gene results in an ls-tim (long) or s-tim (short) isoform of TIM, which exhibit differential sensitivity to light (Peschel et al. 2006). ls-TIM binds poorly with CRY because of an extra 23 amino acids in its N-terminus and hence is less sensitive to light, whereas s-TIM strongly binds with CRY and exhibits more light sensitivity (Peschel et al. 2006; Tauber et al. 2007; Sandrelli et al. 2007).
Temperature is another important external cue that can synchronize the daily behavior of flies. NorpA has been implicated in thermal entrainment but the cells mediating this are largely unknown (Glaser and Stanewsky 2005). Furthermore, a recent paper also showed a role of chordotonal mechanosensory organs in temperature entrainment (Sehadova et al. 2009). These organs are found in peripheral tissues that send signals to the central clock neurons to synchronize daily activity in the presence of temperature cycles. This function is dependent upon expression of the nocte gene in the chordotonal organs (Sehadova et al. 2009). While the focus of this chapter is on mechanisms underlying the control of behavior by central clock neurons, we note that many peripheral clocks can independently entrain to environmental signals. For instance, isolated explants of different tissues entrain to temperature cycles (Glaser and Stanewsky 2005). Similarly, CRY is expressed in peripheral tissues that harbor a bona fide circadian clock such as the Malpighian tubules and eyes, and it may serve to entrain these clocks as well constitute part of the clock mechanism (Ivanchenko et al. 2001; Collins et al. 2006).
3.10 Coupling Between Clock Cell and Their Outputs
There is ample evidence for the existence of several different neuronal subsets and oscillators in the fly brain, but the question remains, how do these groups talk to each other? Several studies have shown the existence of a hierarchical structure in the clock network, based upon alteration of membrane excitability or peptide signaling (reviewed in Nitabach and Taghert 2008). The LNvs are the master pacemakers that send signals, primarily peptidergic (see below), to other clock neurons, but it appears that some of the other neurons also affect the LNvs. The DN1 and DN3 neurons are thought to be glutamatergic and they send projections to the s-LNvs (Hamasaka and Nassel 2006). Manipulating the expression of the glutamate receptor in the LNvs results in alter circadian behavior in DD and substantial effects on the rest–activity pattern in LD (Collins et al. 2012; Hamasaka et al. 2007). The LNvs are also positive for the 5-HT receptor, which affect larval neurophysiology by regulating calcium levels (Hamasaka and Nassel 2006). Altering the 5-HT receptor in the LNvs reduces light entrainment of rest–activity behavior in adult flies (Yuan et al. 2005). Another neurotransmitter gamma-aminobutyric acid (GABA) also alters the output of LNvs by affecting intracellular calcium levels (Hamasaka et al. 2005). In addition, recent studies using genetic, pharmacological, and/or anatomical approaches have also indicated a role for other neurotransmitters such as histamine and acetylcholine (Hamasaka and Nassel 2006; Wegener et al. 2004).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


