Target species
Tissue sample
Systematic namea
Probe sequence (5′–3′)
References
Actinobacillus pleuropneumoniae
Lung, ear, joints, bone, brain
S-S-Ap-0185-a-A-19
CCCACCCTTTAATCCATAG
Genus Brachyspira
Intestine
L-G-Serp-1410-a-A-19
GTCATTCCATCGAAACATA
Brachyspira hyodysenteriae
Intestine
L-S-S.hyo-1210-a-A-19
CTCACGATGAACCTTCGAC
Brachyspira murdochii
Intestine
L-S-B.murdoc-1219-a-A-18
CCGCCTTACGGCAAACCT
[17]
Brachyspira pilosicoli
Intestine
S-S-S.pilo-0209-a-A-18
GCTCATCGTGAAGCGAAA
Clostridium perfringens
Intestine
S-S-Cperfring-0185-a-A-18
TGGTTGAATGATGATGCC
Clostridium difficile
Intestine
S-S-Cl.diff-0193-a-A-18
TGTACTGGCTCACCTTTG
[19]
Chlamydiales
Intestine
S-O-Chls-0523-a-A-18
CCTCCGTATTACCGCAGC
[21]
Chlamydiaceae
Intestine
S-F-Cla-0574-a-A-18
CTTTCCGCCTACACGCCC
[21]
Chlamydia
Intestine
S-G-Chla-0232-a-A-18
TAGCTGATATCACATAGA
[21]
Chlamydophila
Intestine
S-G-Chlph-0583-a-A-18
CTAACTTTCCTTTCCGCC
[21]
Enterococcus faecium
Intestine
L-S-Efaecium-0347-a-A-18
GTGTCTTCCACATTTCGT
[20]
Erysipelothrix rhusiopathiae
Heart
S-S-E.rhusiopathiae-0449-a-A-18
TCCCTCTTCCTATCGTTC
[22]
Escherichia coli
Intestine
L-S-Eco-1531-a-A-21
CACCGTAGTGCCTCGTCATCA
[23]
Klebsiella pneumoniae
Intestine
L-S-Kpneu-1705-a-A-18
TACACACCAGCGTGCCTT
[20]
Leptospira interrogans
Intestine
S-G-Leptospira-1414-a-A-18
CGGGTGCTCCCCACTCAG
[24]
Lawsonia intracellularis
Intestine
S-S-L.intrac-1148-a-A-20
AACCGGAGCAGTCTCTCTAG
Mycoplasma hyopneumoniae
Lung, heart
S-S-M.hyop-0466-a-A-18
CCGTCAAGACTAGAGCAT
[29]
Mycoplasma hyosynoviae
Lung, heart, joints
S-S-M.hyos-0466-a-A-18
CCGTCAGTTCAGTTGCAT
[29]
Mycoplasma hyorhinis
Lung, heart
S-S-M.hyor-0193-a-A-18
GCTGTGAAGCTCCTTTCT
[29]
Pasteurella multocida
Lung, heart, kidney
S-S-Pmul-0449-a-A-20
CTATTTAACAACATCCCTTC
Salmonella spp.
Intestine
L-S-Sal-1713-a-A-18
AATCACTTCACCTACGTG
Streptococcus suis
Heart, brain
S-S-S.suis-0183-a-A-18
CCATGCGGTAAATACTGT
Treponema spp.
Intestine
S-S-Treponema-0833-a-A-18
CCCAGTCCTCATGACCAG
[24]
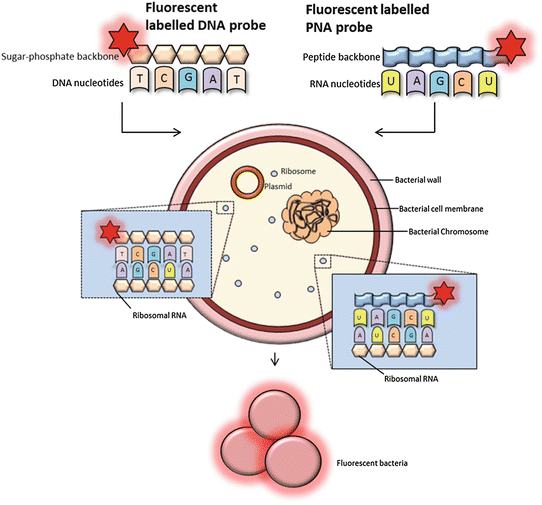
Fig. 1
FISH using oligonucleotide probes. FISH probes can be classified as DNA or PNA probes depending on the structure of the backbone. PNA probes are non-charged which allow a tighter and more specific binding to RNA nucleic acid targets. DNA probes must overcome a destabilizing electrostatic repulsion during hybridization. The fluorescently labeled probes target RNA of the bacterial ribosomes (rRNA). The 16S and 23S rRNA gene sequences contain hyper variable regions that can provide species-specific signature sequences useful for bacterial identification, which most often is utilized in FISH. A adenine, T thymine (DNA specific), G guanine, C cytosine, and U uracil (RNA specific)
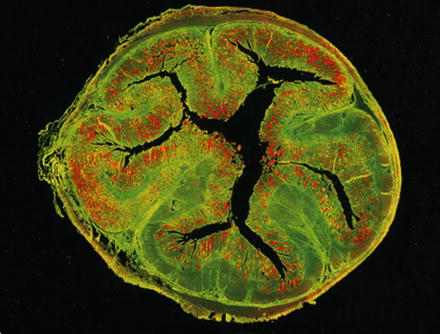
Fig. 2
Visualization of Lawsonia intracellularis (red) by in situ hybridization within a cross section of a porcine ileum. The tissue is green due to autofluorescence. From: Mølbak et al., 2008, Vet Microbiol, 128, 96–107 [25], with permission
The FISH technique consists of four steps: (1) fixation and permeabilization of the sample, (2) hybridization, (3) removing unbound probe, and (4) detection of labeled cells (Figs. 2 and 3). Tissue samples must be fixed and permeabilized prior to hybridization to preserve nucleic acids and ensure that the fluorescently labeled probes can enter the cells. At hybridization, the probe binds to a specific sequence of rRNA inside the bacterial cells (Fig. 1). After removing any unbound probe by briefly rinsing, the tissue sample is ready for screening for reacting cells by fluorescence microscopy [3, 7].
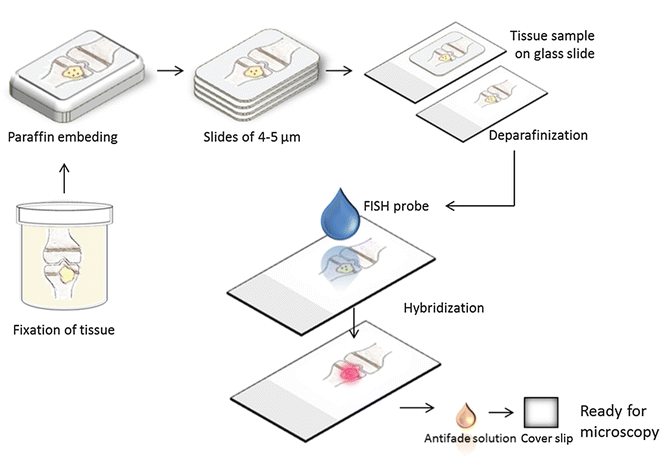

Fig. 3
A bone with osteomyelitis is fixated in formalin and afterward embedded in paraffin. The block of paraffin is cut with a microtome and the tissue is mounted on glass slides. It is necessary to remove the paraffin before the application of the FISH probe. Following hybridization, it is possible to see the fluorescent bacteria within the tissue using a microscope or a scanner equipped for fluorescence
The classical DNA probes consist of fluorescently labeled oligonucleotides or polynucleotides, targeting a specific sequence of microbial rRNA (Fig. 1) [3]. Moreover, peptide nucleic acid (PNA) probes have been developed (Fig. 1). The PNA probes, which are oligomers of single bases linked by a peptide backbone, are more stable to degradation and hybridize to complementary sequences with a higher affinity compared to the classical probes [1, 3, 7]. Probes can be labeled directly or indirectly (Fig. 4). Direct labeling of probes does not require further procedures after hybridization. Indirect labeling of probes uses a reporter molecule or an enzyme bound to the probe and will often result in a brighter signal (Fig. 4). After hybridization, a fluorescent antibody or substrate for enzymatic signal amplification is added depending on the type of labeling of the probe [1]. Identification of different bacterial species in a single tissue sample can be carried out by the application of different fluorophores labeled to each type of probe (Fig. 5).
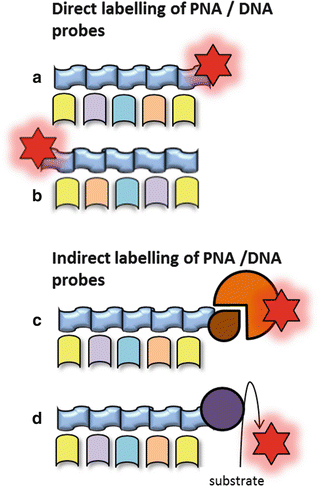
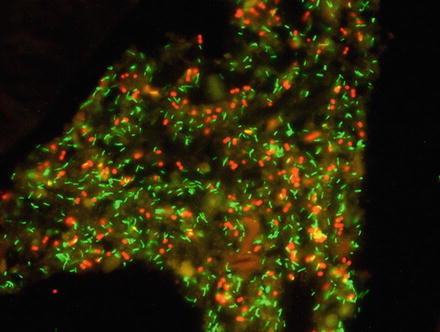

Fig. 4
(a) Direct labeling of the 3′ tail of the probe. (b) Direct labeling of the 5′ tail of the probe. (c) Labeling of the probe with a reporter molecule which is detected by a fluorescent antibody. (d) Labeling of the probe with horseradish peroxidase (HRP) using a substrate for an enzymatic signal amplification

Fig. 5
Visualization of Bifidobacterium spp. (green) and Megasphaera elsdenii (red) by in situ hybridization in a formalin-fixed porcine colon tissue sample
A number of genus- and species-specific probes have successfully been designed and applied for the detection of bacteria of importance in the pig (Table 1), some being available commercially, e.g., Staphylococcus aureus [8].
In the following sections of materials and methods, two different methods (I and II) for FISH are included (Subheadings 2.4 and 3.5 for Method I and Subheadings 2.5 and 3.6 for Method II). Both methods require fixation of tissue (Subheadings 2.1 and 3.1; Fig. 3), deparaffinization (Subheadings 2.2. and 3.2; Fig. 3), and preparation of the probe solution (Subheadings 2.3, 3.3, and 3.4).
2 Materials
2.1 Fixation of Tissue (Fig. 3)
1.
Tissue sample.
2.
10 % neutral-buffered formalin, pH 6.8 (formaldehyde, 37–40 %: 100 mL/L; distilled water 900 mL/L; sodium phosphate, monobasic 4.0 g/L; sodium phosphate, dibasic (anhydrous) 6.5 g/L).
3.
70 %, 96 %, and 99 % ethanol.
4.
Xylene.
5.
Paraffin wax.
6.
Bowl of water.
7.
Plastic container with a tight-fitting lid.
8.
Tissue cool plate.
9.
Microtome.
10.
Adhesive glass slides, e.g., SuperFrost Plus slides (Gerhard Menzel GmbH, Braunschweig, Germany), poly-l-lysine-coated glass (Ted Pella, Inc., Redding, CA, USA), or Biobond Tissue Section Adhesive (AX-LAB, Copenhagen, Denmark).
2.2 Deparaffinization (Fig. 3)
1.
Paraffin-embedded tissue sections on glass slides.
2.
Xylene.
3.
96 % and 99.9 % ethanol.
4.
Sterile water.
5.
Incubator (60 °C).
2.3 Probe Solution
2.
Hybridization buffer, 50 mL of 1 M Tris–HCl (pH 7.2), 90 mL of 5 M NaCl, 5 mL of 10 % (w/v) sodium dodecyl sulfate (SDS), and H2O up to total volume of 500 mL (the final composition of the hybridization buffer is 100 mM of Tris–HCl (pH 7.2), 0.9 M of NaCl, and 0.1 % of SDS).
3.
Crushed ice.
4.
Styrofoam box.
5.
Microcentrifuge tubes (1.5 mL).
2.4 Method I: Hybridization with PNA or DNA Probes on Tissue Sections Mounted on Glass Slides (Fig. 3)
1.
Tissue sections on glass slides (deparaffinized).
2.
Probe solution.
3.
Coverslips, thickness 0.15 mm.
4.
Pencil.
6.
Washing buffer (55 °C), 50 mL of 1 M Tris–HCl (pH 7.2), 90 mL of 5 M NaCl, and H2O up to total volume of 500 mL (the final composition of the washing buffer is 100 mM of Tris–HCl (pH 7.2) and 0.9 M of NaCl) (see Note 1 ).
7.
Antifading mounting media, e. g., VECTASHIELD mounting media with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA, USA) or ImmunoSelect Antifading Mounting Medium (Dianova Gmbh, Hamburg, Germany).
8.
Fluorescence microscope equipped with a 40–100× oil objective and a 100 W mercury lamp or similar, camera for fluorescence, and the correct filter sets for the chosen fluorescent-labeled probes to be used.
9.
Nonfluorescent immersion oil for microscopy.
2.5 Method II: Hybridization with DNA or PNA Probes on Tissue Sections Mounted on Glass Slides Using a Shandon Rack (Fig. 6)
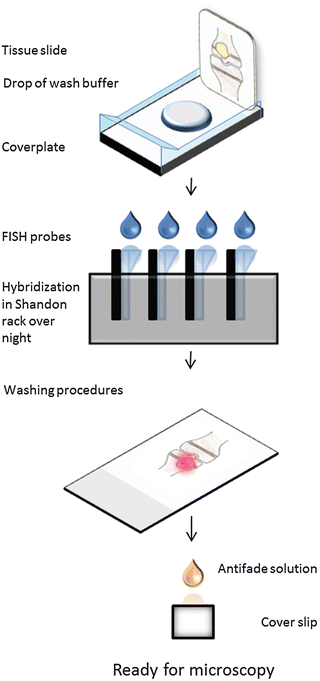
Fig. 6
Detection of bacteria with FISH in tissue mounted on glass slide using a Shandon rack
1.
Tissue sections on glass slides (deparaffinized).



