Filoviridae
Abstract
This chapter describes the properties of filoviruses and features of the diseases they cause in animals.
Keywords
Filovirus; Marburgvirus; Ebolavirus; Filoviridae; Rousettus aegyptiacus; Egyptian rousette bat
In 1967, 31 cases of a severe hemorrhagic fever, with 7 deaths, occurred among laboratory workers in Germany and Yugoslavia who were processing kidneys from African green monkeys (Cercopithecus aethiops) that had been imported from Uganda. A new virus isolated from the tissues of these patients and monkeys was named Marburg virus, now the prototype member of the genus Marburgvirus in the family Filoviridae. Nine years later, in 1976, two separate epidemics of hemorrhagic fever with high mortality occurred, one in villages in the rain forest of Zaire (now the Democratic Republic of Congo) and the other in southern Sudan (now Republic of South Sudan). Viruses that were morphologically identical to, but antigenically distinct from, Marburg virus were isolated and named Ebola virus (genus Ebolavirus). Later, the viruses from Zaire and Sudan were found to be genetically distinct and are now designated Ebola virus and Sudan virus. Since 1976, two more species of ebolavirus were discovered in Africa, Taï Forest virus in the Ivory Coast in 1994, and Bundibugyo virus in Uganda in 2007. Collectively, these ebolaviruses and marburgviruses have caused more than 25 sporadic epidemics in Africa of Ebola virus disease and Marburg virus disease, formally designated as Ebola and Marburg hemorrhagic fevers. In 1989 and 1990, monkeys imported from the Philippines into a quarantine facility in Reston, Virginia, were infected with a fifth type of ebolavirus species, now called Reston virus. Infected monkeys at the facility became ill, and many died. Four animal caretakers were infected, but developed no clinically apparent disease. Reston virus was subsequently identified in monkeys imported from the Philippines into Italy (1992) and Texas (1996) and then in farmed pigs and their handlers in the Philippines in 2008. Lastly, in 2012, an ebolavirus-like filovirus was reported in dead bats (Miniopterus schreibersii) in Europe. This virus has been named Lloviu virus (genus Cuevavirus).
The member viruses of the family Filoviridae are intriguing for several reasons: (1) the viruses, although similar to other members of the order Mononegavirales (the viruses comprising the families Paramyxoviridae, Rhabdoviridae, Filoviridae, and Bornaviridae, see Chapter 17: Paramyxoviridae and Pneumoviridae) in their genomic organization and mode of replication, are morphologically the most bizarre of all viruses; (2) the viruses have caused large disease outbreaks and therefore are recognized as having substantial epidemic potential, as evidenced by the recent (2013-2016) Ebola virus disease outbreak in West Africa in which more than 28,000 people were infected with more than 11,000 deaths; (3) the viruses cause a devastating clinical disease in humans and nonhuman primates, including chimpanzees, gorillas, and macaques, with extremely rapid and florid tissue damage, and with high mortality; (4) although these infections are clearly zoonotic, much remains to be established regarding their epidemiology, but recent evidence indicates that species of fruit bats may be reservoir hosts of both ebola- and marburgviruses. The viruses that cause these diseases are Biosafety Level 4 pathogens and classified as Tier 1 Select Agents; they must be handled in the laboratory under maximum containment conditions to prevent human exposure.
Properties of FILOVIRUSES
Classification
All nonsegmented negative-sense RNA viruses share several characteristics: (1) a similar genome organization and roughly the same gene order; (2) a virion-associated RNA polymerase (transcriptase); (3) a helical nucleocapsid; (4) transcription of messenger RNAs (mRNAs) by sequential interrupted synthesis from a single promoter; (5) virion maturation via budding of preassembled nucleocapsids from the cellular plasma membrane at sites containing patches of viral glycoprotein spikes (peplomers). These characteristics and conserved ancient domains found in genomic nucleotide sequences support the notion of a common ancestry, as reflected in the establishment of the order Mononegavirales. Conserved domains in the nucleoprotein and polymerase genes indicate that viruses in the family Filoviridae are related most closely to those in the genus Orthopneumovirus in the family Pneumoviridae, rather than to viruses in the family Rhabdoviridae as might be expected from their similar, helically wound nucleocapsid structures.
The family Filoviridae contains three genera, Marburgvirus, consisting of a single species Marburg marburgvirus that has two genetically distinct viruses isolated in Kenya, namely Marburg virus and Ravn virus; Ebolavirus, with five recognized species of Sudan ebolavirus (Sudan virus), Zaire ebolavirus (Ebola virus), Reston ebolavirus (Reston virus), Bundibugyo ebolavirus (Bundibugyo virus), and Taï Forest ebolavirus (Taï Forest virus); and Cuevavirus that contains a single uncultured virus species Lloviu cuevavirus (Lloviu virus) that was detected in dead cave-dwelling bats in Spain (Fig. 19.1).
Virion Properties
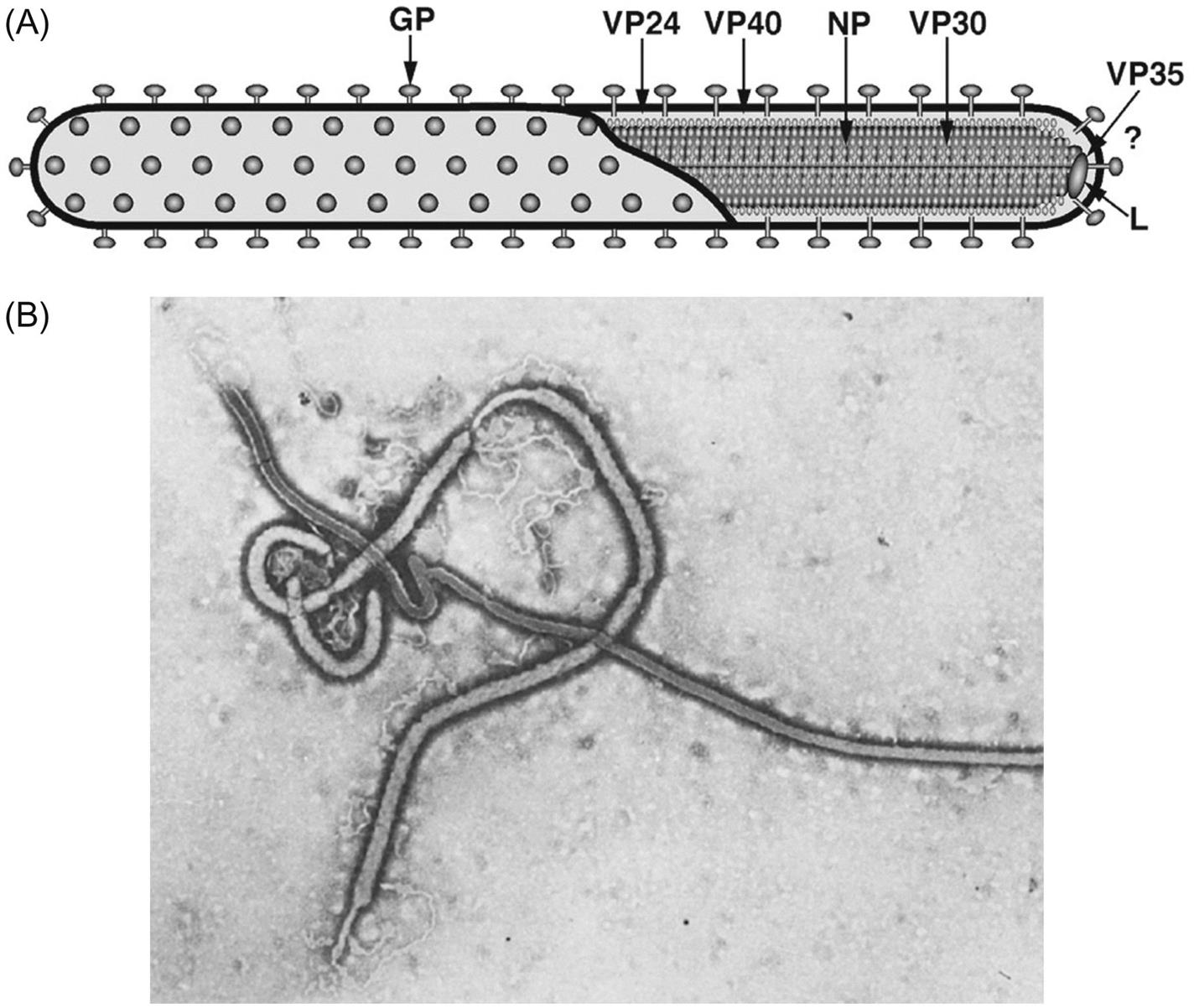
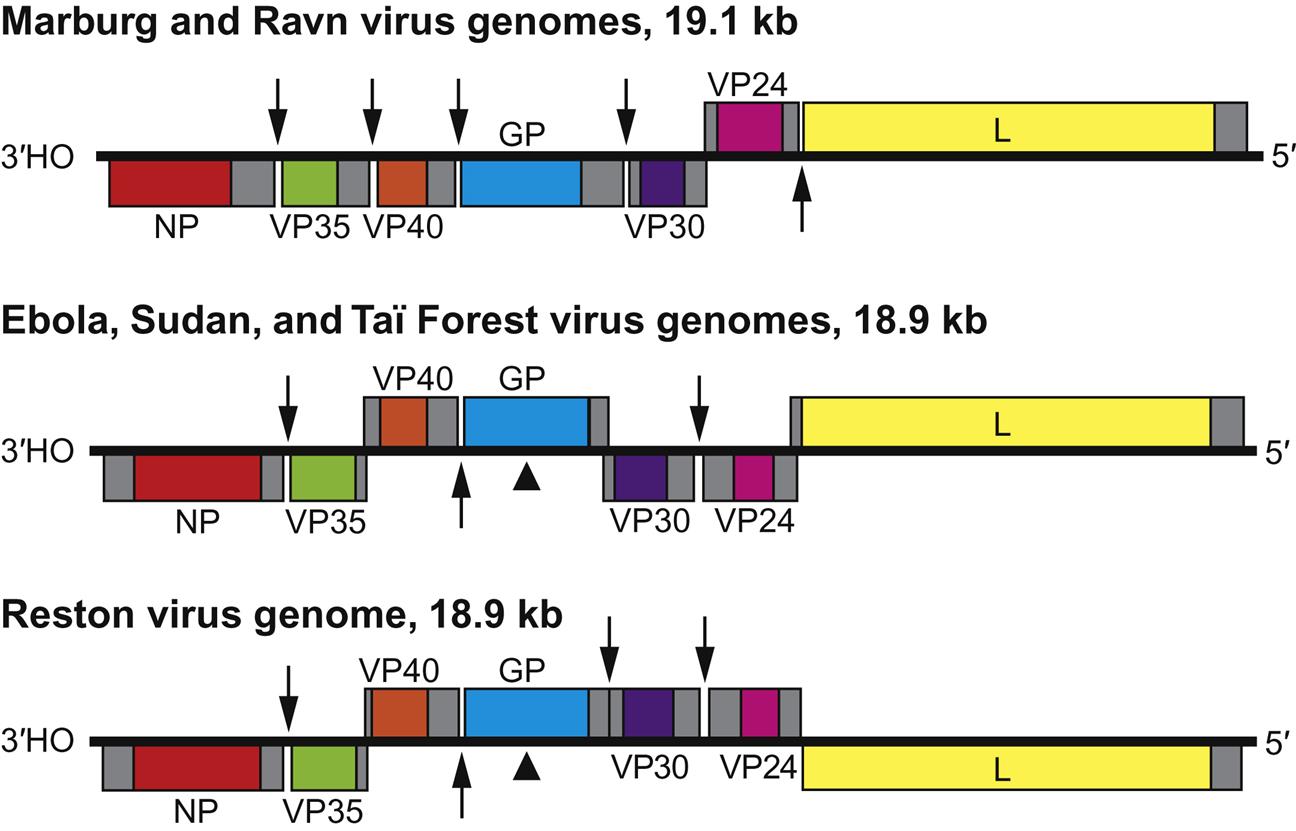
Filovirus virions are markedly pleomorphic, appearing as long, filamentous, sometimes branched forms, or as U-shaped, “6”-shaped, or circular forms. Virions have a uniform diameter of 80 nm and vary greatly in length (particles may be up to 14,000 nm long, but have unit nucleocapsid lengths of about 800 nm in the case of marburgvirus and 1000 nm for ebolavirus). Virions are composed of a lipid envelope covered with trimeric glycoprotein spikes, surrounding a helically wound nucleocapsid (50 nm in diameter). They include at least seven proteins, including the RNA-dependent RNA transcriptase/polymerase (L), the surface glycoprotein (GP), the nucleoprotein (NP), the matrix protein (VP40), a phosphoprotein equivalent (VP35), a polymerase cofactor (VP30), and a membrane-associated protein (VP24) (Fig. 19.2; Table 19.1). GP consists of two domains, GP1, a receptor binding subunit, and GP2, a membrane fusion unit. The genome is a single molecule of negative-sense, single-stranded RNA, approximately 19 kb in size, the largest of all negative-sense RNA viruses. The gene order is: 3′-NP–VP35–VP40–GP–VP30–VP24–L-5′ (Fig. 19.3). Genes are separated either by variable intergenic sequences or by overlaps—that is, short (17–20 bases) regions where the transcription start signal of the downstream gene precedes the transcription stop signal of the upstream gene. Ebolaviruses have three overlaps that alternate with intergenic sequences, whereas marburgviruses have a single overlap (Fig. 19.3).
Table 19.1
Virions are pleomorphic, appearing as long filamentous forms and other shapes; they have a uniform diameter of 80 nm and vary greatly in length (unit nucleocapsid lengths of about 800 nm for Marburg and 1000 nm for Ebola virus)
Virions are composed of a lipid envelope covered with spikes surrounding a helically wound nucleocapsid
The genome is composed of a single molecule of negative-sense, single-stranded RNA, 19.1 kb in size
Infection is extremely cytopathic in cultured cells and in target organs of host
Cytoplasmic replication, large intracytoplasmic inclusion bodies, budding from the plasma membrane
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


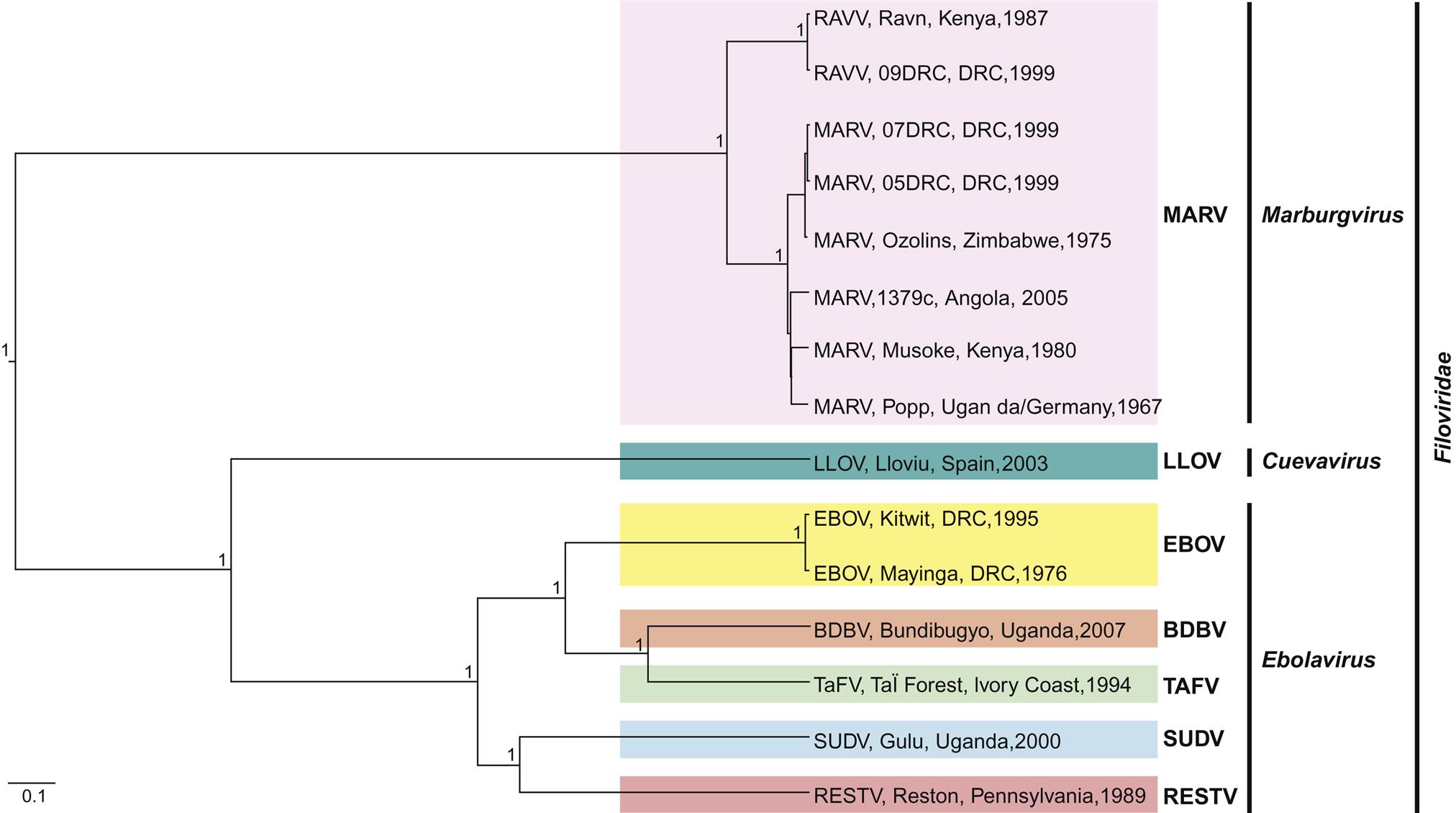
 Forest virus; SUDV, Sudan virus; RESTV, Reston virus.
Forest virus; SUDV, Sudan virus; RESTV, Reston virus.