Retroviridae
Abstract
This chapter describes the properties of retroviruses and features of the diseases they cause in animals.
Keywords
Retroviruses; oncogene; alpharetrovirus; betareovirus; lentivirus
Retroviruses infect a wide variety of animal species including mammals, reptiles, birds, and fish; these viruses are associated with many economically important diseases. Retrovirus infections are typically confined to a single host species (host-adapted) and rarely cross species barriers. Diseases induced by retroviruses are diverse and, depending on the individual virus and its associated disease, can include distinct pathogenic mechanisms involving inflammation, neurodegeneration, immunodeficiency, and cellular transformation (neoplasia). All retroviruses are enveloped, single-stranded, positive-sense RNA viruses with a duplicated (diploid) genome that requires a novel reverse transcription step during their replication cycle. During reverse transcription, the viral RNA genome is converted into a double-stranded DNA copy, a seeming violation of a central dogma of biology. Reverse transcription is accomplished by the virus-encoded enzyme reverse transcriptase, discovered in 1970 by Howard Temin, Renato Dulbecco, and David Baltimore, all of whom subsequently received the Nobel Prize. It is difficult to overstate the importance of this discovery, as it has facilitated seminal advances in diverse fields of biology including virology, molecular genetics, cellular growth/carcinogenesis, and diagnostic medicine.
Diseases associated with retrovirus infections have been recognized for more than a century. Equine infectious anemia, Jaagsiekte (pulmonary adenomatosis) of sheep, and bovine leukosis were all described in the 19th century, long before their causative etiology was understood. Retroviruses in tissue filtrates from chickens with leukosis were investigated by physician Vilhem Ellerman and veterinarian Oluf Bang in Copenhagen in 1908. They were able to transmit leukemia by inoculating chickens with cell-free filtrates. Peyton Rous, a medical pathologist, succeeded in producing transplantable sarcomas in chickens by injecting chicken tumor-derived cell-free filtrates in 1910–1911 (the eponymously named Rous sarcoma virus). Nearly 60 years after this discovery, Rous was awarded the Nobel Prize.
Properties of RETROVIRUSES
Key biological features of retroviruses include the ability to undergo mutation and recombination, acquire and alter host-derived genetic sequences, covalently integrate into the host-cell genome, and activate or inactivate specific host genes near the site of their integration. These features of the biology of retroviruses have been exploited by researchers to generate vectors that deliver nonviral transgenes (gene therapy). In recent years, many of the details of the biology of retroviruses have been gleaned from intense research focused on the human immunodeficiency virus (HIV).
Classification
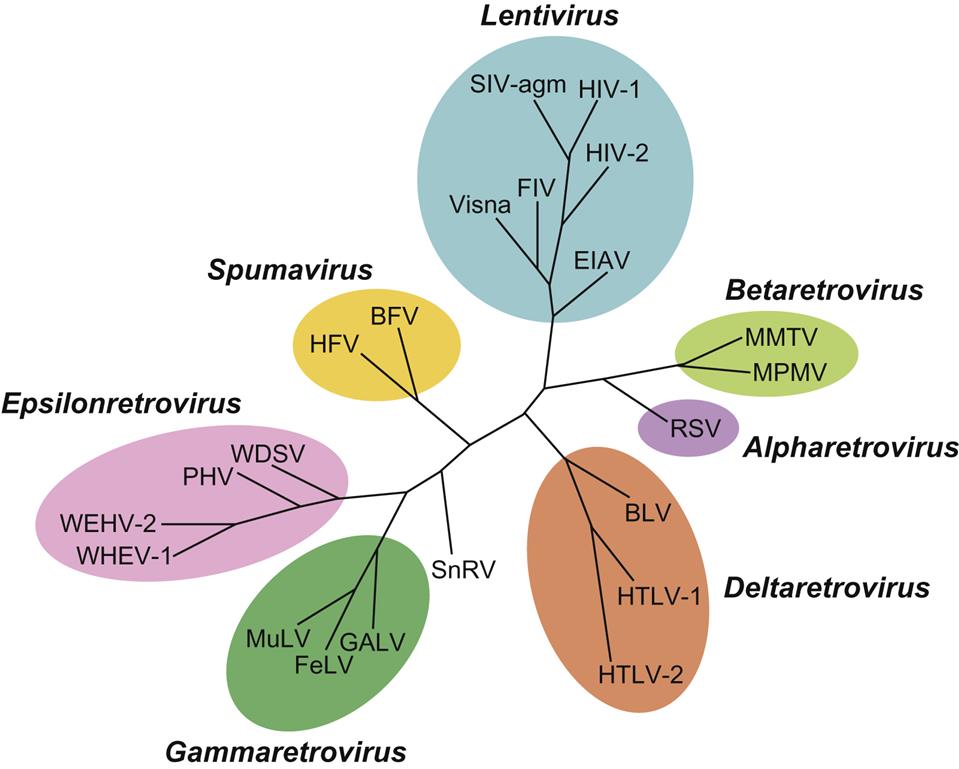
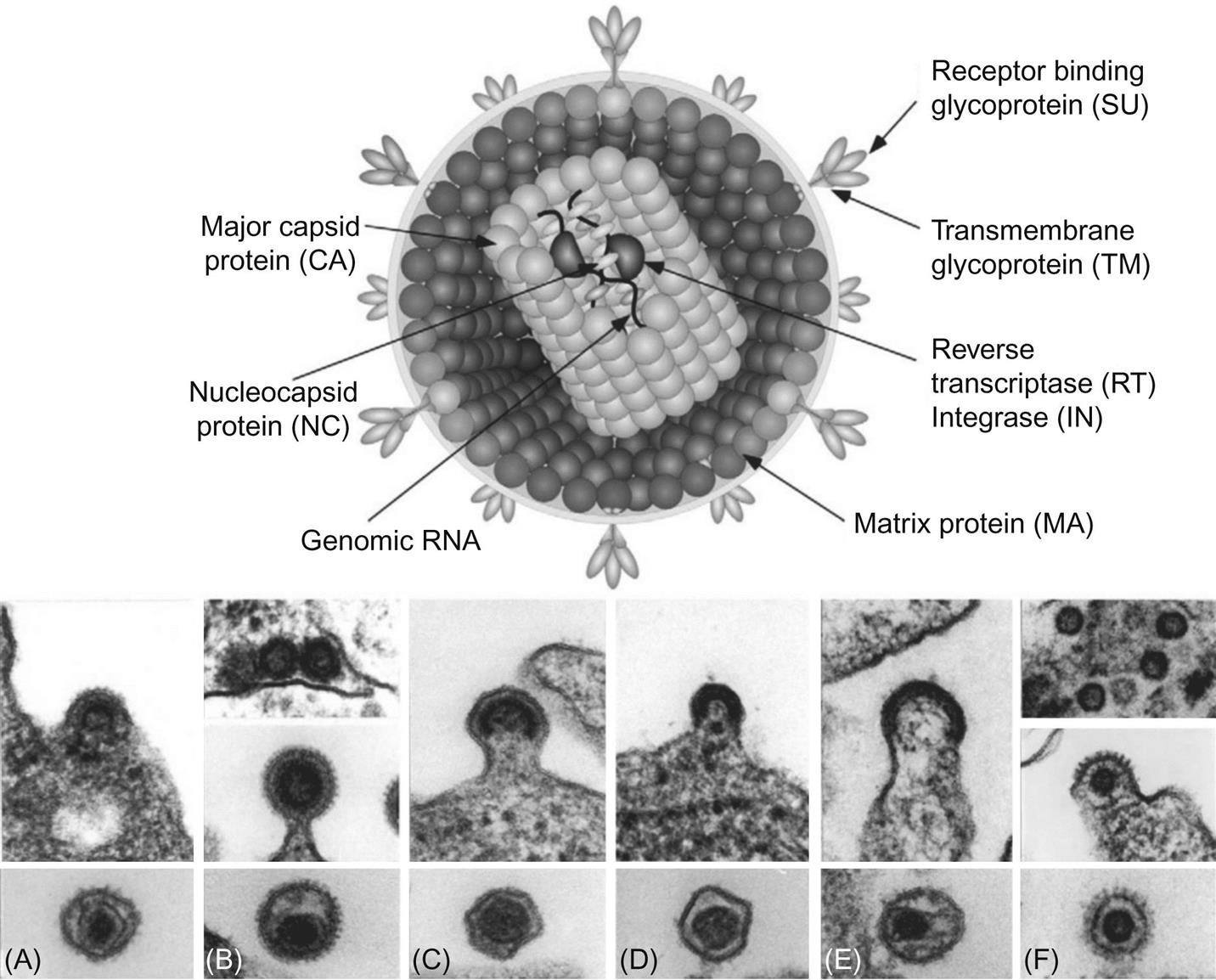
The family Retroviridae (Latin retro, “backward”) is divided into two subfamilies (Orthoretrovirinae and Spumaretrovirinae) and seven genera (Fig. 14.1). Classification of retroviruses is complicated by an older classification scheme based upon the ultrastructural appearance of the virion. In this system, still in use, four different types of virus particles are recognized and designated as A, B, C, and D-type retroviruses. The morphology of virions is classified into structures with: an intracellular double membrane lacking budding forms (A), an extracellular eccentric, spherical core (B), a central, spherical core (C), or a cylindrical core (D) (Fig. 14.2). Other classification schemes are based upon horizontal versus vertical transmission of individual retroviruses, or their ability to transform infected cells (oncogenic retroviruses). Currently, the most widely accepted taxonomic classification of retroviruses is based upon their genomic sequence; viruses are thereby grouped by evolutionary relatedness (Fig. 14.1).
The subfamily Orthoretrovirinae is further subdivided into six genera, Alpharetrovirus, Betaretrovirus, Gammaretrovirus, Deltaretrovirus, Epsilonretrovirus, and Lentivirus, whereas the subfamily Spumaretrovirinae includes only a single genus, Spumavirus (Fig. 14.1). Viruses included in five of these genera are potentially oncogenic (formerly known as oncoviruses), whereas members included in the genera Lentivirus and Spumavirus are not oncogenic. Lentivirus infections are characterized by a prolonged incubation period between initial infection to occurrence of disease (from the Latin “lenti,” meaning “slow”). Spumaviruses are named for their tendency to cause distinctive and characteristic vacuolation of the cytoplasm of infected cells (from the Greek “spuma,” meaning “foam”). Despite significant efforts, spumaviruses have not been convincingly associated with diseases in animals.
Virion Properties
Retrovirus virions are 80–100 nm in diameter with a genome that includes four main genes encoding the virion proteins: gag, pro, pol, and env. The gag gene, or group-associated antigen, encodes the major structural nonglycosylated polyproteins: specifically, matrix (MA), capsid (CA), and nucleocapsid (NC) (Figs. 14.2 and 14.3). The pro gene encodes a protease responsible for facilitating viral protein maturation, while the pol gene encodes the multifunctional protein that includes the reverse transcriptase (RT) and integrase enzyme functions. The env gene encodes the antigenic surface glycoproteins (SU) and transmembrane protein (TM).
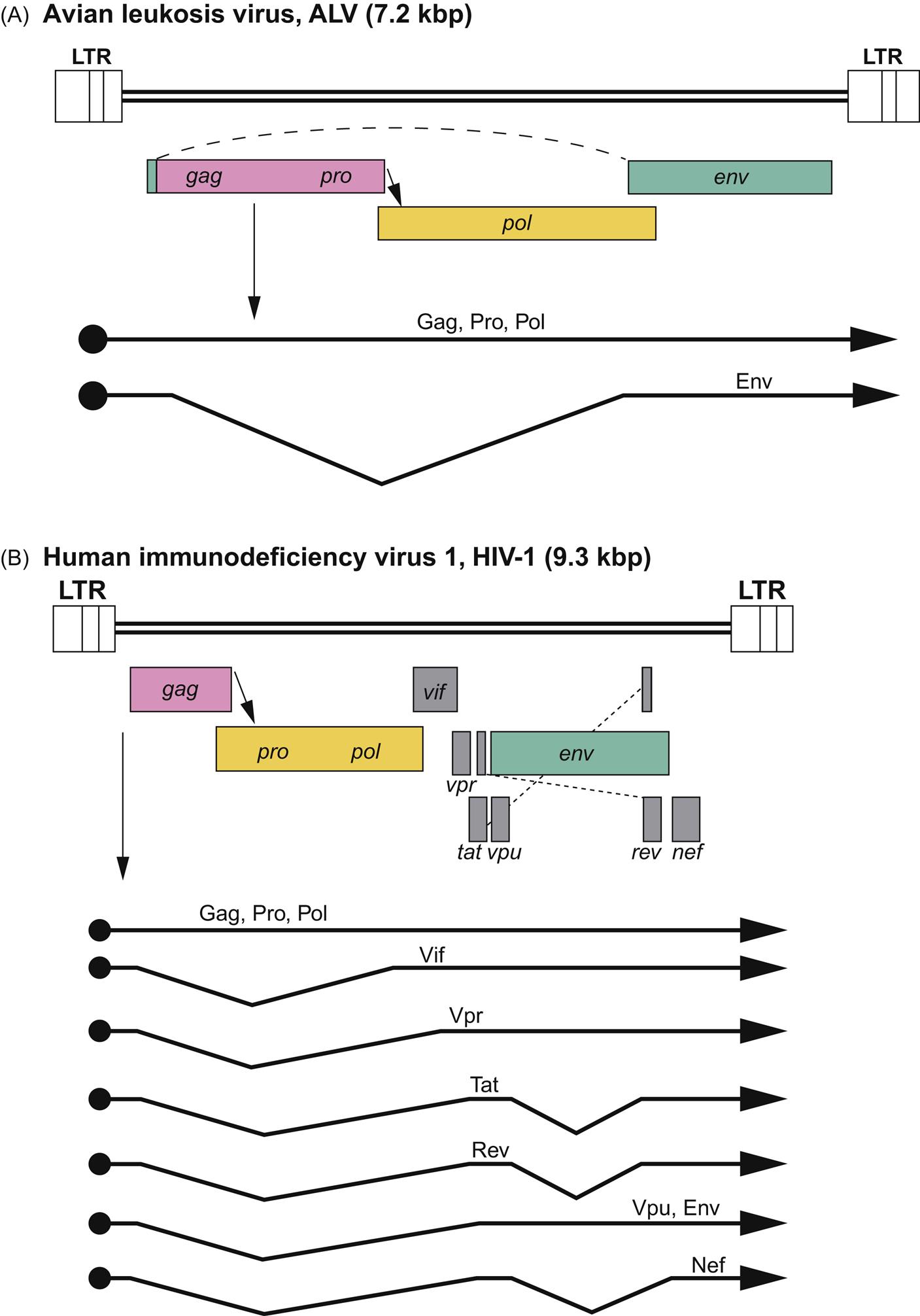
The genome of retroviruses is unique in being comprised of two 7–12 kbp copies of positive-sense RNA that makes retroviruses effectively diploid. The two hydrogen-bonded monomers have a short dimerized region, a 5′ CAP and an approximately 200 nucleotide 3′ poly-A tail. Genome termini have identical repeat (R) regions near both ends and unique U5 and U3 regions at the 5′ and 3′ ends, respectively. These and adjacent regions are critical for initiation of reverse transcription, integration, and postintegration transcription. Although the standard retrovirus genome is depicted as 5′-R U5 gag pro pol env U3 R-3′, the more complex retroviruses (such as deltaretroviruses, epsilonretroviruses and lentiviruses) include additional genes that are important for the regulation of gene expression, virus replication, and pathogenesis (Fig. 14.3).
Retrovirus virions have a three-layered structure comprised of the various Gag polyproteins (Fig. 14.2). At the virion core are two copies of the virus genome complexed with NC proteins in a helical structure. This core structure is enclosed within a polymer of CA proteins, forming the icosahedral capsid. Polymerized MA proteins surround the capsid, which in turn is surrounded by the host-derived lipid envelope. The lipid envelope is studded with the SU and TM glycoproteins that are responsible for receptor binding and membrane fusion. Antibodies that neutralize virus infectivity in cell culture are often directed towards the envelope glycoproteins, although such antibodies are not always predictive of neutralization in vivo because of the emergence of so-called neutralization resistant “escape mutant” variant viruses. The presence of the lipid envelope allows inactivation by lipid solvents, detergents, or heat. Retroviruses are, however, more resistant than many other viruses to ultraviolet and X-irradiation, in part because their diploid genomes compensate for radiation-induced mutations during reverse transcription.
Since the reverse transcriptase enzyme is essential for conversion of the genomic RNA into provirus DNA (shortly after virus entry into a susceptible cell), approximately 30 copies of this enzyme are packaged into the virion core. The reverse transcriptase requires either a Mg2+ or Mn+2+ cation for activity, and most retroviruses utilize Mg2+. The protein product of the pol gene has multiple functions, and can variously serve as an RNA-dependent DNA polymerase (reverse transcriptase), a DNA-dependent DNA polymerase, and an RNase. Each of these enzymatic functions is carried out by a different part of the protein molecule. Reverse transcriptase is more error prone than the host cell DNA polymerases due to its lack of a 3′–5′ exonuclease proofreading mechanism. The introduction of sequence errors (mutations) during the RNA → DNA reverse transcription process plays a critical role in establishing genetic diversity of retroviruses within the infected host. Recombination is another important mechanism of generating genetic diversity in retroviruses. As a consequence of frequent template switching between the two monomers of the diploid genome by the viral reverse transcriptase, cells infected with two related but genetically different retroviruses can produce heterozygous virions. Stable genetic recombinant viruses are then generated in cells infected by these heterozygous virions. The generation of virus sequence diversity is a key feature of retrovirus persistence, evolution, and pathogenesis.
Virus Replication
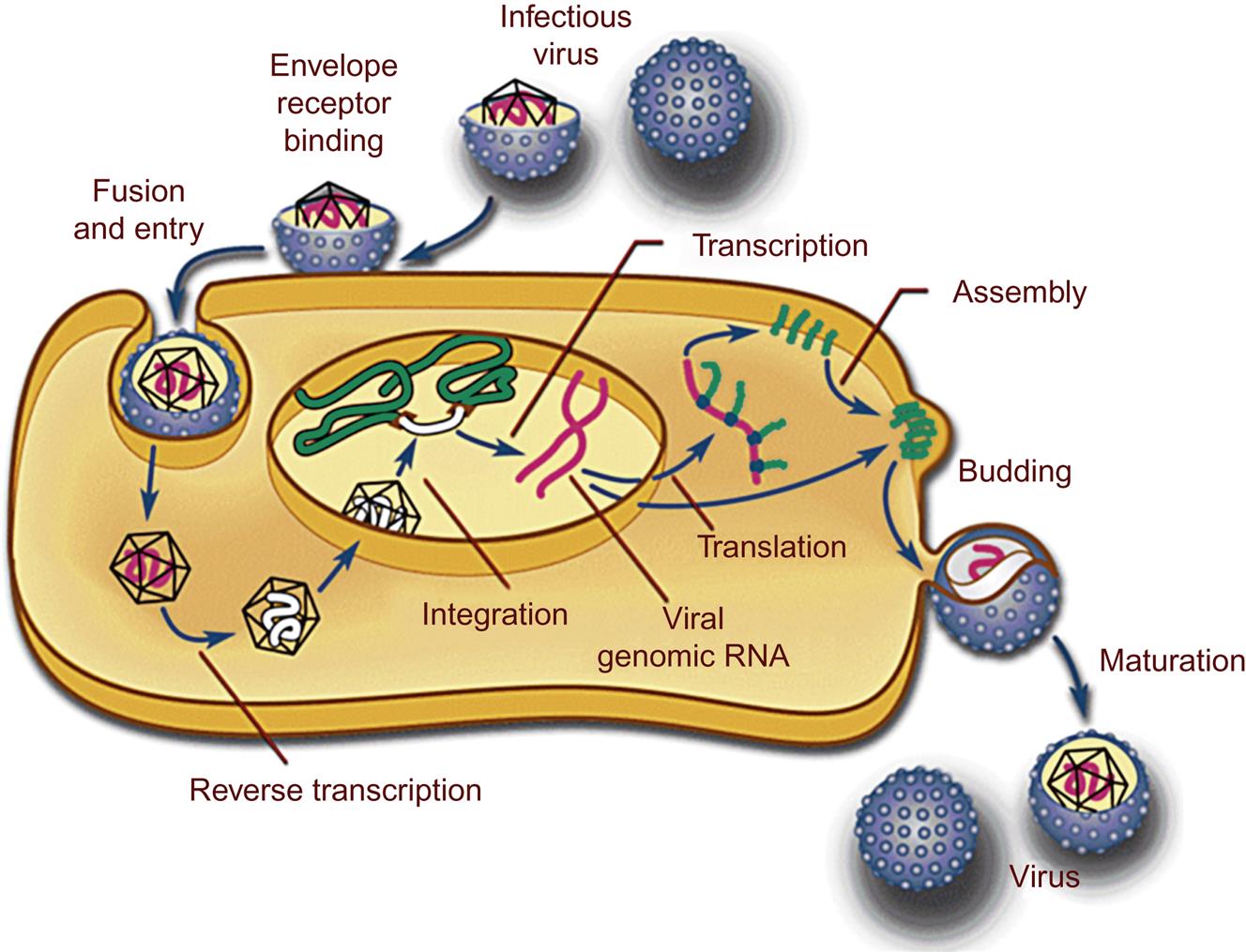
As described in detail in Chapter 2: Virus Replication (Fig. 2.10), the replication cycle of retroviruses begins with engagement of the virus’ envelope glycoprotein surface receptor with cell-surface ligands (Fig. 14.4). This highly specific and molecularly complex step effectively restricts the host range of the virus. The specificity of the cell-surface receptors defines both the susceptibility of host species as well as cell tropism. For example, immunodeficiency-inducing lentiviruses such as feline immunodeficiency virus and human immunodeficiency virus are able to bind, enter, and infect both monocyte/macrophages and CD4+ T cells of their respective hosts (cats and humans), whereas an inflammation-inducing lentivirus such as caprine arthritis encephalitis virus will infect only goat monocyte/macrophages and not CD4+ T cells. Virus attachment is followed by virus–host cell membrane fusion, facilitating insertion of the virus core into the cell cytoplasm. Less commonly, cell entry is mediated by endocytosis. The core complex of retroviruses consists of the diploid retrovirus genome, reverse transcriptase and integrase enzymes, and associated Gag proteins. Following penetration of the virus core particle into the cytoplasm, the strategically error-prone reverse transcriptase enzyme synthesizes a DNA copy of the virus’ single-stranded, positive sense (+) RNA genome through a complex molecular mechanism. A host cell-derived transfer RNA (tRNA) serves as an RNA primer for the reverse transcriptase enzyme by binding near the U5 region of the viral RNA. Each retrovirus has a particular tRNA for optimized priming. The multifunctional reverse transcriptase enzyme synthesizes a complementary strand of R-U5 DNA to form a negative (–) strand DNA/positive (+) strand RNA hybrid at the 5′ end of the viral genome.
The infectious genomic RNA of retroviruses lacks the flanking long terminal repeat (LTR) regions of the double-stranded DNA copy, or provirus. The identical LTRs are synthesized through a complex replication mechanism involving two “jumps” of the reverse transcriptase enzyme between the 5′ and 3′ template molecules. The key to understanding this replication step is the R region of the virus’ genome; the R region is a direct repeat located near both genomic termini. After reverse transcriptase synthesizes a segment of negative (–) strand DNA complementary to the 5′ R region, restricted digestion of the template RNA occurs (from the RNase function of the reverse transcriptase protein) to expose the single-stranded DNA. The exposed single-stranded DNA then forms a DNA-RNA hybrid with either the 3′ R region of the same template molecule (intramolecular) or the 3′ R region of the second strand of the diploid genome (intermolecular). Either way, the formation of this DNA–RNA hybrid creates a template (first jump) for the RT enzyme to complete reverse transcription of the retrovirus genome to the 5′ R-U5. To complete the U3-R-U5 LTR, the 5′ U3 region is copied from the completed 3′ LTR (second jump). The result is a complete viral genome with paired terminal LTRs (provirus).
Reverse transcriptase-mediated synthesis of the provirus DNA occurs in the cell cytoplasm. The provirus is subsequently translocated into the nucleus where the integrase enzyme facilitates integration of the linear provirus copy into the host cell genome. Most retroviruses (except lentiviruses) rely on cell division for efficient passage into the nucleus. Integration into the host chromosomal DNA is not entirely random, as it tends to occur in regions of open chromatin (areas of the chromosome undergoing active transcription). Once the provirus is integrated into the host genome, it is relatively stable (genetically) and can be replicated and transcribed by cellular mechanisms utilizing host DNA and RNA polymerases. As the viral promoter, the 5′ LTR functions as a form of “processor,” evaluating a variety of cellular and environmental signals to optimize the timing and magnitude of virus replication. This region also contains multiple enhancer elements, the ribosome binding site (start of translation), transcription termination signals, and a signal for the addition of the 3′ poly (A) tail. The provirus may remain latent (transcriptionally inactive) or alternatively, actively transcribe viral RNA to varying degrees of efficiency (transcriptionally active). The signaling mechanisms and epigenetic regulation of retrovirus transcription is an active field of investigation germane to both retrovirus pathogenesis and gene therapy technology.
Transcription of the integrated provirus results in both full-length viral RNA molecules (genomic RNA) and mRNA molecules that have been spliced into smaller RNAs by the host cell spliceosome to provide template for the Gag, Pro, Pol, Env, and the accessory viral proteins. In simple retroviruses, transcription and splicing are accomplished by host cell molecules, whereas in complex retroviruses (members of the genera Deltaretrovirus and Lentivirus), in conjunction with host cell proteins, virus-encoded proteins like Tat and Rev directly and dramatically influence viral transcription and RNA splicing. The viral RNA molecules are transported into the cytoplasm and are translated into viral proteins by host cell ribosomes. In many retroviruses, effective ribosomal scanning of viral mRNA is dependent upon frame-shift events to produce all of the required gene products. These frame-shift events allow a single string of genetic material in the virus’ genome to encode multiple gene products. The Env proteins SU and TM are glycosylated as they are processed in the cytoplasmic Golgi apparatus; these proteins are subsequently trafficked to the plasma membrane.
Virions assemble at the plasma membrane (for the majority of genera), or as intracytoplasmic particles (betaretroviruses and spumaviruses). Full-length genomic viral RNA is “packaged” with Gag proteins to form the virus core structure. In most retroviruses, a critical stretch of nucleotides known as the packaging signal is present between the 5′ LTR and the gag open reading frame (5′ untranslated region or 5′ UTR). Newly formed virions are released from the cell surface by “budding” through specialized membrane microdomains known as lipid rafts (see Fig. 2.12). During budding, the virions are coated with an outer sphere of plasma membrane-derived lipid molecules containing the embedded envelope glycoproteins. Notably, completion of the retrovirus replication (life) cycle usually does not lead to death of the host cell (cell lysis).
RETROVIRUS-Induced Oncogenesis
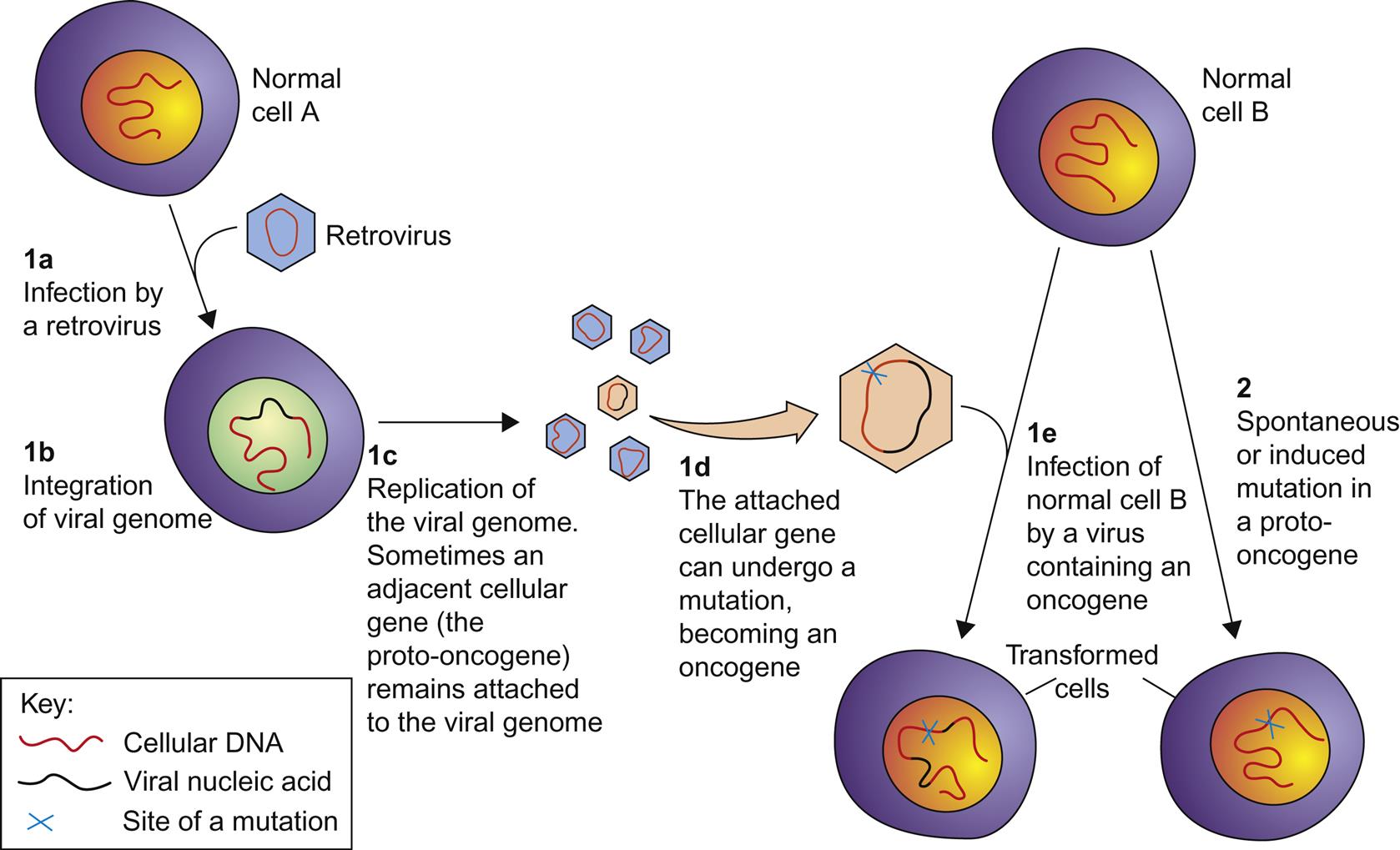
Oncogenic retroviruses infect and transform cells in all classes of vertebrates, but especially well characterized examples occur in chickens, inbred mice, ruminant livestock, and domestic cats. Research into virus-induced oncogenesis has been pursued for more than a century and this research has uncovered many of the complex details governing cellular growth. Tumor-inducing genes (oncogenes) were discovered through the study of retroviruses and associated animal tumors. In 1976, Stehelin, Varmus, Bishop, and Vogt discovered the src oncogene in Rous sarcoma virus, which was responsible for virus-induced cell transformation. In 1989, Bishop and Varmus were awarded the Nobel Prize for this seminal discovery. Retroviruses are capable of transforming cells in at least three different ways: (1) the process of insertional mutagenesis; (2) through “capture” and unregulated expression of a host cell proto-oncogene; and (3) certain retroviruses, eg, bovine leukemia virus, Jaagsiekte sheep retrovirus, and enzootic nasal tumor virus, include genes that encode protein products that are directly oncogenic to infected cells.
Insertional Mutagenesis
The insertion of the provirus into the host cell genome is, by definition, mutagenic. Retroviruses often have transcriptionally powerful promoters, potentially influencing the expression of neighboring host genes at the site of insertion. If a host gene near the provirus insertion site happens to be a proto-oncogene, cellular transformation may result. Proto-oncogenes are host genes that regulate cell growth; as such they encode growth factors, growth factor receptors, intracellular signaling molecules, or transcription factors. As proto-oncogenes, these important genes are normally under stringent cellular control. Retrovirus-induced insertional mutagenesis disrupts this tight regulation, potentially resulting in unregulated expression of the proto-oncogene and cellular transformation. Less commonly, integration of provirus can also disrupt a tumor suppressor gene, leading to loss of this gene product and resultant cellular transformation.
Oncogene Capture
Retroviruses can capture a host cell proto-oncogene, incorporate it into their genome, and subsequently pass it on to progeny viruses that aberrantly express the gene in an unregulated manner (Fig. 14.5). Such aberrant expression can result in cellular transformation; a gene expressed in this manner is referred to as a viral oncogene (v-onc). Viruses that express their own oncogene are designated as strongly (or rapidly) transforming. Proto-oncogene capture can occur as a result of transcript read-through into the adjacent host DNA, and subsequent recombination events incorporate the host gene into the viral genome. Mutations may also occur within the captured oncogene during virus replication, resulting in constitutive unregulated function when the viral oncogene is expressed. The captured cellular oncogene typically replaces some of the virus’ own genetic material, resulting in a replication-incompetent (syn. replication-defective) virus. In order to replicate, such replication-defective viruses must infect a cell that is coinfected with another intact virus (referred to as a helper virus) that can provide the missing viral gene product(s) in trans. The alpharetrovirus Rous sarcoma virus is a noted exception to this rule; it has a full complement of the expected retrovirus genes in addition to a v-onc gene. As a result, this virus is replication-competent without the need for a coinfection with a helper virus, and infection with Rous sarcoma virus also results in rapid oncogenesis. The transduction of cellular proto-oncogenes has only been found in the simple retroviruses, and has not been identified in complex retroviruses such as the lentiviruses. Why this is so is not yet clear.
Oncogenic Retrovirus Genes
The third mechanism of viral oncogenesis is mediated through the products of viral genes that are themselves oncogenic. This mechanism is limited to viruses in certain genera, notably the Deltaretrovirus and Betaretrovirus. For example, the env gene product of the sheep betaretroviruses, specifically enzootic nasal tumor virus and Jaagsiekte sheep retrovirus, is itself oncogenic, and infection of sheep with these viruses can result in the production of nasal or pulmonary carcinomas, respectively. This fascinating pathogenesis has been demonstrated through elegant transgene expression experiments in mouse models.
Endogenous RETROVIRUSES
Through time, integration of retroviruses into the host cell genome has provided both an opportunity and a mechanism for retroviruses to infect and be perpetuated within the germline tissues of virtually all species of animals. Infection/insertion of retroviruses into the germline allows for their vertical transmission between individuals, with the integrated provirus inherited in a Mendelian fashion (so-called “genetic transmission”). Genome sequencing has shown that a significant proportion of animal genomes are comprised of transposable genetic elements, or transposons; a substantial subset of these elements is in fact comprised of endogenous retroviruses. For example, sheep have approximately 27 copies of endogenous retroviruses stably integrated into their genomes, and these endogenous retroviruses are closely related to the “exogenous” betaretroviruses of sheep, namely Jaagsiekte sheep retrovirus and enzootic nasal tumor virus. Many (or most) endogenous retroviruses have multiple premature stop codons and other mutations sprinkled throughout the provirus genome. As a result, endogenous retroviruses are typically incapable of producing infectious virus particles, except in inbred mice. Controversy exists as to whether exogenous retroviruses have evolved from endogenous retroviruses, or alternatively, whether endogenous retroviruses are the evolutionary remnants of a prior infection event with an exogenous retrovirus.
Once thought to be inert, it is now evident that endogenous retroviruses can influence the cells in which they are found. In respect to cellular pathology, both positive and negative roles have been proposed for endogenous retroviruses via their potential role in homologous recombination-induced chromosomal rearrangement between distant loci, and their direct influence on gene expression. Endogenous retroviruses also act as transposons, which integrate randomly during cell division, thereby creating de novo mutations. Many mouse phenotypes (strain characteristics) are due to such events, for example retinal degeneration and hair coat color. There is also compelling evidence that some endogenous retroviruses interact and interfere with pathogenic exogenous retroviruses—those transmitted between animals as typical virus infections—thereby protecting the host cell from infection. Still, the net effect of the endogenous retrovirus “cargo” on their host cells (and the association of endogenous retroviruses with disease) remains controversial and somewhat enigmatic.
Members of the Subfamily Orthoretrovirinae
As the biological behavior of individual viruses included in the subfamily Orthoretrovirinae is reflective of the general properties of the different genera in the subfamily, the various orthoretroviruses will be described according to their taxonomic grouping (by genus) rather than by the species of animal they infect.
Genus ALPHARETROVIRUS
MEMBERS OF THE GENUS AVIAN LEUKOSIS, SARCOMA, and Related ALPHARETROVIRUSES (AVIAN LEUKOSIS COMPLEX VIRUSES)
The genus Alpharetrovirus includes many important avian viruses; avian leukosis/sarcoma virus is the genus prototype. These viruses exhibit “C-type” morphology, and have a simple genomic organization (Fig. 14.3). The study of alpharetrovirus infections in poultry (leukosis/sarcoma group or historically called avian leukosis complex viruses) has played a critical role in the emergence of virology as a discipline. In the first part of the 20th century, infections with avian leukosis virus caused great economic losses to the poultry industry. As a result, alpharetroviruses were intensively studied by animal scientists and virologists motivated by these interesting and economically important pathogens. Tumors resulting from avian leukosis/sarcoma virus infections were the first virus-induced tumors to be identified in any species. In many instances, these avian tumors resulted from the aberrant expression of retrovirus oncogenes; the modern-day molecular study of viral oncogenes and viruses of the avian leucosis/sarcoma group has revolutionized our understanding of oncogenesis (syn. carcinogenesis, tumorigenesis).
Clinical Features and Epidemiology
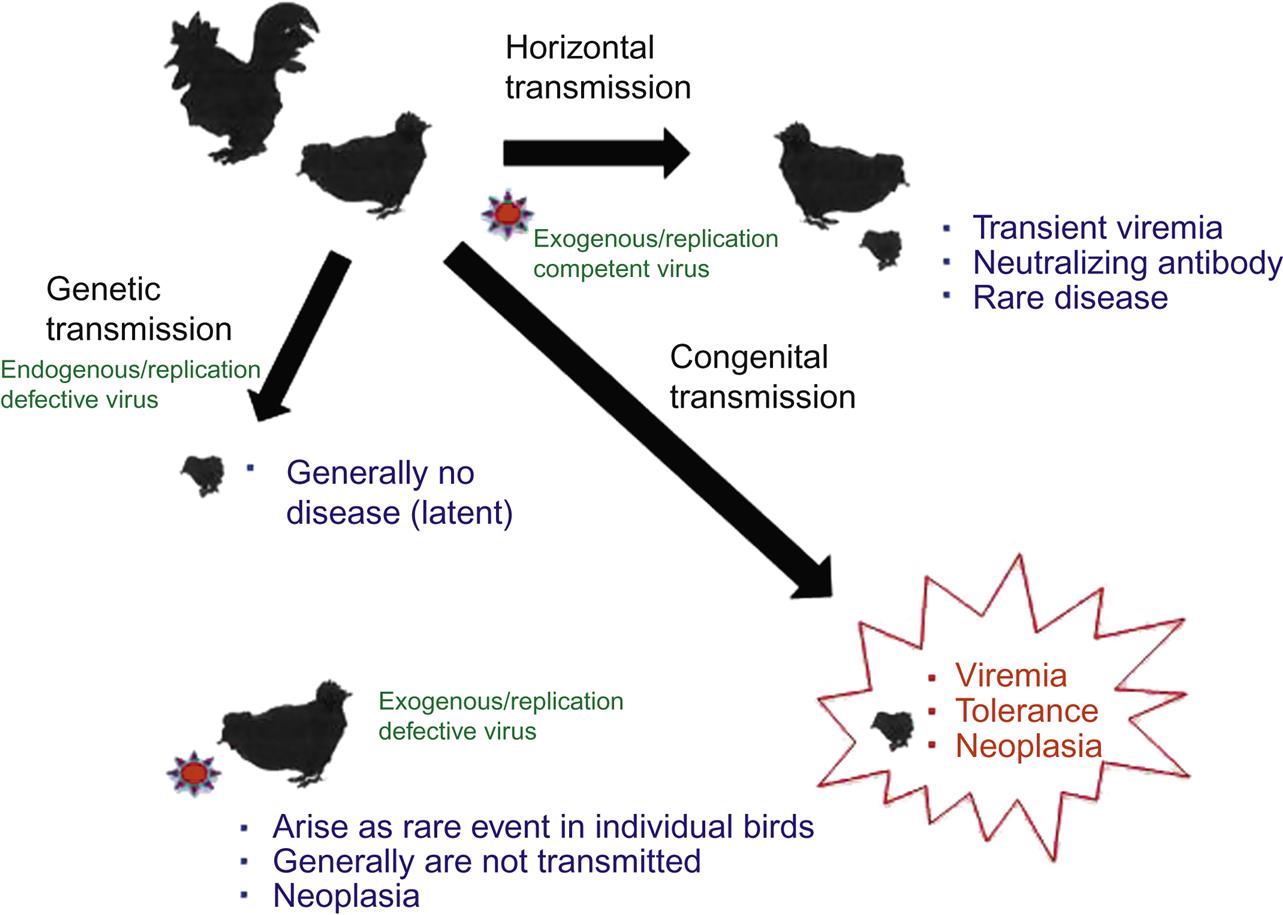
The alpharetroviruses are genomically simple viruses that can be transmitted from bird to bird in three different ways, namely horizontally, congenitally, and genetically. The varied mechanisms of transmission of these agents have important consequences for pathogen control. Horizontal transmission occurs from an infected bird to another, either directly or through infected fomites (Fig. 14.6). Horizontal infection is often, but not always, nonpathogenic. Most birds develop a transient viremia that is effectively controlled and eliminated by neutralizing antibody. However, a few infected birds may eventually develop leukemia or lymphoma. In contrast, congenital infection occurs when an infected hen transmits the retrovirus from her oviduct to her developing egg. Infected males do not appear to infect their progeny. As a result of immunologic tolerance, many congenitally infected birds develop a persistent viremia with no obvious antibody response. Such birds shed large amounts of virus into the environment and can subsequently develop various neoplastic conditions. Lastly, genetic transmission occurs when an integrated endogenous retrovirus is transmitted in Mendelian fashion from infected gamete to chick. Such a transmission event (which occurs in most birds) usually results in a silent (latent) infection and generally does not result in either viremia or tumor formation (neoplasia).
The alpharetroviruses include a substantial number of related but distinct viruses (avian leukosis/sarcoma viruses) that can be: (1) exogenous (meaning these are transmitted between birds as typical viruses) and replication-competent; (2) exogenous but replication-defective; or (3) endogenous and replication-defective viruses. The exogenous/replication-competent viruses can be transmitted horizontally or congenitally and have the standard complement of gag, pol, pro, and env genes (Fig. 14.3). The rate of tumor development among birds infected with such viruses, if any, is generally slow because these viruses do not carry their own oncogene and any tumors they induce arise from insertional mutagenesis. One of these viruses, however, Rous sarcoma virus, has the standard complement of retrovirus genes plus a viral oncogene (ie, it is an exogenous, replication-competent and oncogene-containing alpharetrovirus). As a result, Rous sarcoma virus is both replication-competent and rapidly/acutely transforming. In contrast, the other avian alpharetroviruses that have captured an oncogene have then sacrificed one of their own genes in exchange for the captured one (meaning that they have lost one or more of their complement of gag, pro, pol, or env genes). Such viruses are exogenous but replication-defective; to complete their life cycle, they require the presence of a helper virus to provide the missing viral gene products in trans. Although they are replication-defective, such viruses are often rapidly transforming as a result of the captured oncogene. In general, these defective viruses arise as rare events in each individual bird in which they are found; because they are replication-defective and are so rapidly fatal, they are rarely transmitted from bird to bird.
Endogenous avian leukosis viruses were discovered in the 1960s when researchers identified retroviral gag and env gene products inherited as if they were dominant autosomal genes in chick embryos that were free of infectious avian leukosis virus. Subsequently, scientists demonstrated that nearly all chicken somatic and germline cells contain partial to complete avian leukosis virus sequences integrated into their genomes. The endogenous avian leukosis viruses have an overall genetic structure similar to exogenous avian leukosis viruses, but most also have numerous mutations (stop mutations/deletions) that prevent expression and assembly of infectious virus. Transmission of endogenous avian leukosis virus from parent to chick is via genetic transmission.
Viruses in the avian leukosis complex are divided into at least 10 subgroups, 6 of which infect chickens (A, B C, D, E, and J). This classification scheme is based upon variation in the surface envelope glycoprotein, which determines the virus-serum neutralization properties and cellular tropism of individual viruses. Incidence of infection varies greatly by country with rare occurrence of infection in genetic stocks of commercial poultry in the United States because of the testing and elimination program of the National Poultry Improvement Plan. However, in many other countries, most commercial layer flocks of chickens will be infected with avian leukosis/sarcoma virus within the first few months of hatching. Historically, disease occurs only sporadically (~1–2% incidence) in birds over 14 weeks old if rapidly transforming viruses like Rous sarcoma virus are not present. In the 1990s, however, when avian leukosis virus J was first detected in the United States in broiler stocks, the incidence of disease soared to 20%. Today, however, commercial layer and broiler breeding industries have nearly eradicated avian leukosis/sarcoma virus from primary breeding flocks through selection of resistant breeds and testing and elimination of infected birds, and the occurrence of avian leukosis/sarcoma in the field is generally from horizontal transmission of the virus amongst commercial layer birds in the United States and is not common.
Pathogenesis and Pathology
At the beginning of the 20th century, Ellerman identified and characterized multiple strains of avian leukosis virus and created a classification scheme to define the various neoplasms and leukemias they induced, specifically as to their cellular origin as erythroid, myeloid, or lymphoid. This classification scheme is still employed today.
Lymphoid Leukosis
In 1981, Hayward and colleagues found that lymphomas (syn. lymphosarcoma or malignant lymphoma) of the cloacal bursa that were caused by exogenous/replication-competent avian leukosis virus, were associated with provirus integration adjacent to the cellular c-myc proto-oncogene. Integration of the 3′ LTR of the provirus apparently induced transcriptional expression of the adjacent cellular proto-oncogene c-myc. These bursal lymphomas (originating in B cell lymphoblasts) took time to develop (slowly transforming virus), often originating from a background of lymphoid hyperplasia. Such a pathogenesis is consistent with the concept of insertional mutagenesis. Lymphoid leukosis, synonymous with visceral lymphomatosis, is the most common disease associated with avian leukosis virus infection and occurs in chickens that are 14–30 weeks of age. Clinical signs are often nonspecific, but the comb may be pale and occasionally cyanotic. Affected birds may be inappetent, emaciated, weak, and have a distended coelomic cavity. Neoplasms may be present for some time prior to the onset of clinical signs, and they may only be recognized at the time of slaughter of aged laying or breeding chickens when their presence typically results in carcass condemnation. The disease is rare in young broilers. Lymphoid neoplasms are typically multifocal, discrete nodular/proliferative masses located in the cloacal bursa, liver, spleen, and other viscera. Interestingly, a bursectomy, even up to 5 months of age, will block the development of lymphoid leukosis.
Mesenchymal Tumors and Osteopetrosis
Avian leukosis virus infections in poultry can also induce mesenchymal neoplasms, osteopetrosis, or renal neoplasms. Infection with particular alpharetroviruses can result in a variety of solid mesenchymal neoplasms, including fibrosarcoma, fibroma, myxosarcoma, myxoma, histiocytic sarcoma, osteoma, osteosarcoma, and chondrosarcoma. These neoplasms are the result of infections with exogenous retroviruses containing v-onc genes, either replication-competent viruses such as Rous sarcoma virus or replication-defective viruses. Osteopetrosis (so-called thick leg syndrome) is most often a uniform or irregular diaphyseal thickening of the long bones of the leg. Affected birds may also have anemia and lymphoid leukosis. Renal neoplasms include nephroblastomas and renal carcinomas. Both renal tumors and osteopetrosis are the result of infections of birds with exogenous/replication-competent avian leukosis complex viruses.
Myeloid/Erythroid Tumors
Neoplastic transformation of bone marrow derived erythroid (red blood cells) or myeloid (granulocytic) cells occurs in birds infected with exogenous/replication-defective alpharetroviruses that include a v-onc gene. Specific types of neoplasm are often associated with aberrant expression of specific oncogenes. For example, v-myc expression is associated with myelocytomatosis (myeloid cell leukemia), v-myb expression is associated with myeloblastosis (myeloblast leukemia), and aberrant v-erbA or v-erbB expression is associated with erythroblastosis (erythroid cell leukemia). At least 15 different oncogenes have been identified in avian alpharetroviruses.
Diagnosis
History, clinical signs, gross lesions, and histopathology are generally sufficient to diagnose avian leukosis. For differential diagnosis, however, it is to be stressed that two additional avian viruses can also induce lymphoma, namely Marek’s disease virus (a herpesvirus, see Chapter 9: Herpesvirales) and avian reticuloendotheliosis virus (a gammaretrovirus that also induces B cell lymphomas; see later in this chapter).
Replication-competent acutely transforming viruses like Rous sarcoma virus can be quantitated using proliferative focus formation assays in chick embryo fibroblast cell cultures. Replication-competent avian leukosis viruses other than Rous sarcoma virus do not transform cells in vitro. However, since such viruses interfere with viruses that do carry a v-onc gene, interference assays in cell culture are often used to quantitate them. Replication-defective rapidly transforming viruses, which carry a v-onc gene, can also be quantified by focus formation in cell culture, but virions will not be produced unless the cultures are coinfected with a replication-competent helper avian leukosis virus.
Serologic detection methods, such as ELISA, can also be used to confirm the presence of avian leukosis virus in a poultry flock, however, it is to be stressed that congenitally infected birds do not develop a detectable immune response. RT PCR assay, using either viral RNA or DNA as template, can be used to detect and quantify most avian alpharetroviruses, but to perform such an assay, one needs to have an index of suspicion of which virus is being assayed (for appropriate primer design). More sophisticated sequence analysis assays (next generation sequencing, deep sequencing; see Chapter 5: Laboratory Diagnosis of Viral Infections) can be employed to identify new virus subgroups associated with unusual disease patterns.
Immunity, Prevention, and Control
Chicks congenitally infected with avian leukosis virus shed the virus in their meconium and feces after hatching and are often avian leukosis virus seronegative as a result of immunologic tolerance. Congenital infection is generally a consequence of virus being present within the oviduct during egg formation in viremic hens. In this scenario, high titers of virus are present in the egg albumin with subsequent embryo infection very early in gestation. Although virus titers in the congenitally infected chick may reach 109 virions/mL of blood, immunotolerant, persistently infected chicks often appear clinically normal. However, a subset of the persistently infected chicks will eventually develop leukosis. These chicks are an important potential reservoir of virus for the flock and are able to transmit virus horizontally in feces and saliva to other birds by close contact. Infected viremic birds transmit virus either continuously (generally viremic, seronegative birds) or intermittently (viremic, seropositive birds). Transmission efficiency decreases in birds older than 18 months of age.
The scenario is different for chicks that have not been exposed to avian leukosis virus congenitally. The efficiency of passive transfer of virus-specific antibody from hen to chick is low; most 1-day-old chicks have maternal antibody titers of just 1–10% of their dam’s titers. By 4–7 weeks of age, most chicks are seronegative. If the chick is horizontally infected at this time, it generally becomes transiently viremic, mounts an effective immune response eliminating the virus, and is seropositive from then on. A few chicks remain persistently infected, serving as a source of virus for the flock as congenitally infected birds do.
Hygiene is important in minimizing environmental contamination, particularly in the posthatching period when age, population density, and levels of virus are all conducive to horizontal transmission. As a result, “all-in-all-out” management systems, associated with thorough disinfection of incubators, hatcheries, brooding houses, and other equipment are standard practice.
Genetic lines of chickens that are genetically susceptible to avian leukosis virus were unwittingly selected when intensive methods for broiler and egg production were introduced in the 1940s. Today, most commercial flocks consist of genetically resistant lines of birds, which has resulted in a sharp reduction in the incidence of avian leukosis. Since genetic resistance of chickens to infection is associated with the lack of specific cell membrane receptor molecules for the alpharetrovirus envelope glycoproteins, it is possible to select for resistant lines by challenging chorioallantoic membrane or chick fibroblast cultures derived from prospective genetic bird lineages with pseudotyped Rous sarcoma virus. Failure to produce foci of cell transformation in vitro indicates genetic resistance, and lines of leukosis-resistant chickens can then be bred. Since viruses with compensatory mutations emerge continuously, genetic resistance as a basis for control of avian leukosis virus requires a continuous selection program.
It is possible to create flocks of birds free of horizontally transmitted avian leukosis. Such flocks are important when the eggs they produce are used for vaccine production. Although these birds are free of exogenous avian leukosis virus, they still carry integrated endogenous avian retroviruses.
Immunization strategies using either live-attenuated or inactivated vaccines have not been successful in preventing disease.
Genus BETAREOVIRUS
The genus Betaretrovirus includes important pathogens of sheep, goats, primates, and laboratory mice. Mouse mammary tumor virus is the genus prototype. Viruses in this genus include both exogenous and endogenous retroviruses of mice, primates, and small ruminants (sheep and goats). No oncogene-containing betaretrovirus has been described, however the murine and small ruminant viruses are associated with different types of neoplasia whereas the primate viruses are associated with immunodeficiency syndromes.
MEMBERS OF THE GENUS JAAGSIEKTE SHEEP RETROVIRUS (OVINE PULMONARY ADENOCARCINOMA VIRUS)
Jaagsiekte sheep retrovirus, also called ovine pulmonary carcinoma virus, is an oncogenic, exogenous retrovirus of sheep and less often goats that causes infectious pulmonary adenocarcinomas/adenomas. The associated disease syndrome is named ovine pulmonary adenomatosis, with synonyms that include ovine pulmonary carcinoma or sheep pulmonary adenomatosis. Jaagsiekte sheep retrovirus has a global distribution with the notable exceptions of Australia and New Zealand. It was eradicated from Iceland in 1952. The disease occurs sporadically in North America and Europe, whereas in Peru, it may be responsible for up to a quarter of the annual mortality in adult sheep.
Clinical Features and Epidemiology
South African sheep farmers originally described ovine pulmonary adenomatosis in the early 1800s, where it was referred to as Jaagsiekte. “Jaagsiekte” is an Afrikaans term that is translated as “chase sickness,” aptly describing the disease syndrome in affected sheep—affected sheep exhibit chronic wasting and have numerous pulmonary adenocarcinomas/adenomas leading to respiratory distress, especially when the animals are stressed through exercise such as herding.
The virus is transmitted horizontally between sheep in close contact by aerosolized pulmonary fluid. Following exposure, there is an extended incubation period of perhaps 1–3 years so that affected sheep are typically adult animals >2 years of age. Experimentally, however, the virus can cause lung tumors in very young lambs. Only a subset of virus-infected sheep ever develop pulmonary adenomatosis, perhaps 30%. Clinically affected sheep have progressive dyspnea, bouts of spasmodic coughing, weight loss, and anorexia. The clinical signs reflect both replacement of pulmonary parenchyma with tumor tissue as well as the copious production of surfactant-containing fluid by the neoplastic cells. The end result is respiratory system failure and, frequently, secondary pneumonia.
Pathogenesis and Pathology
Jaagsiekte sheep retrovirus is a simple retrovirus that does not include a viral oncogene (v-onc). In a series of elegant experiments, the virus’ Env protein was itself shown to induce cellular transformation in vitro and to create pulmonary adenocarcinomas in mice. The virus’ LTR region and env gene product collectively define its tissue tropism. The cellular receptor is hyaluronidase 2 (Hyal2), which is expressed on many cell types and serves as the ligand for the Env glycoprotein. The LTR is transcriptionally active in two specific cell types that are confined to the lung, alveolar type II pneumocytes, and bronchiolar Clara cells, thereby defining the highly restricted tissue tropism of this virus; production of virus proteins has been demonstrated only in pulmonary epithelium, tumor cells, and tumor-associated fluid. Although virus transcription is apparently confined to the lung, integrated provirus DNA is present in lung, lymphoid tissue, alveolar macrophages, and peripheral blood mononuclear cells of infected sheep.
Pulmonary neoplasms, which manifest as solid tumor masses, are often detected at slaughter of culled sheep from endemic flocks, but metastases to the pulmonary lymph nodes are rare. Histologically, pulmonary neoplasms are classified as either adenomas or low-grade adenocarcinomas that are histologically similar to human bronchioloalveolar carcinomas.
Diagnosis
The clinical signs of pulmonary adenomatosis are characteristic, notably the abundant fluid that is produced in the lungs of affected sheep. However, definitive diagnosis requires differentiation from chronic pneumonia caused by the ovine progressive pneumonia lentivirus and by various bacteria, indeed coinfections with these various agents are all common. A unique feature of Jaagsiekte sheep retrovirus infection is that it elicits no obvious antibody response, perhaps because of immune tolerance induced by the presence of closely related endogenous retroviruses in the sheep genome. The lack of any serologic screening assay complicates early detection of infection of sheep. However, PCR-mediated amplification of provirus contained in peripheral blood cells has been used successfully as a screening tool in disease control programs as provirus DNA is present in peripheral blood cells prior to the development of pulmonary tumors.
Immunity, Prevention, and Control
Outbreaks of ovine pulmonary adenomatosis occur typically when virus-infected sheep are introduced into uninfected flocks, especially if the sheep are closely confined. Vaccines are not available, nor would they be effective given the apparent immune tolerance of sheep to the virus. The incidence of disease can be reduced greatly by strict isolation of flocks and the prompt removal of clinically sick animals as they shed large amounts of infectious virus. Eradication of Jaagsiekte sheep retrovirus from Iceland involved the near depopulation of all Icelandic sheep as no screening diagnostic test was available at the time. Countries, regions, and individual flocks that are free of the virus can maintain their status through stringent quarantine and screening of any imported sheep.
MEMBERS OF THE GENUS ENZOOTIC NASAL TUMOR VIRUS
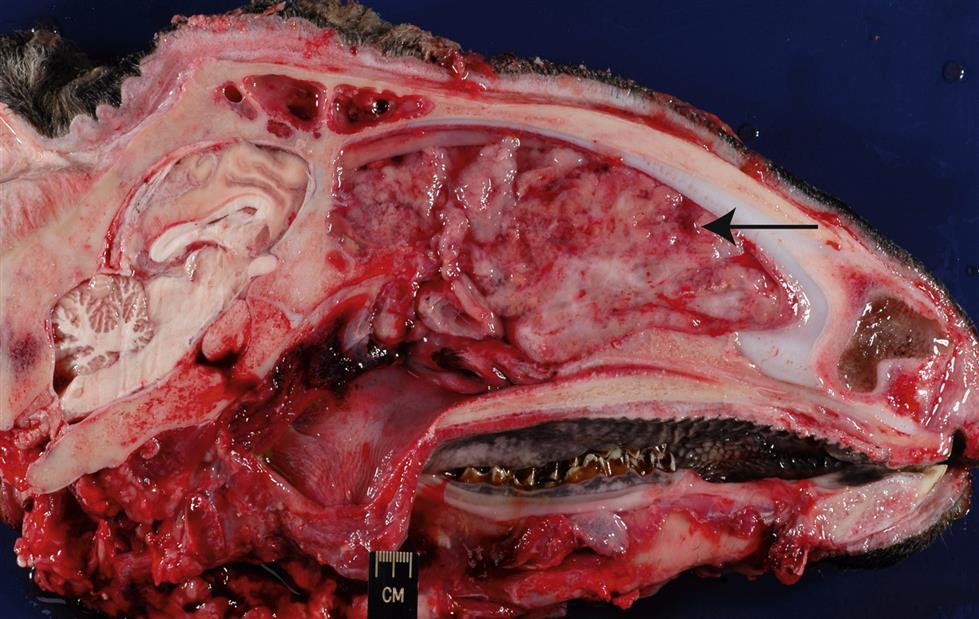
The enzootic nasal tumor virus (ENTV) infects both sheep (ENTV-1) and goats (ENTV-2), and these (ENTV-1 and ENTV-2) are apparently distinct viruses that share ~95% overall amino acid similarity to Jaagsiekte sheep retrovirus. Less is known regarding the global distribution, pathogenesis, and molecular constitution of enzootic nasal tumor virus than the related virus of sheep. The enzootic nasal tumor virus can transform secretory epithelial cells of the ethmoid turbinate, a restricted region of the nasal cavity, thus infections with this virus frequently result in nasal adenocarcinomas (enzootic nasal adenocarcinoma, enzootic nasal tumor; Fig. 14.7). The condition has been experimentally reproduced in lambs following inoculation with enzootic nasal tumor virus.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree