Fig. 1
Schematic structure of a CRISPR/Cas region in MTBC strains and application of the CRISPR array, to the development of the spoligotyping method, using 43 selected spacer sequences. (a) General structure of a CRISPR/Cas region; (b) structure of DR region: 0–56 copies of 36 bp direct repeats, separated by variable spacer sequences of 36–51 bp, whose order is well conserved in the genome of MTBC. Spacers, here numbered from 4 to 8. The DRs together with the adjacent spacers form the direct variant repeats (DVRs); (c) PCR amplification of the DR flanking spacers, using primers DRa and DRb, one of them biotin labeled, complementary to the conserved direct repeats; (d) five pretend patterns in the DR region and possible formation of strain III pattern by insertion of IS6110 sequence in DVR 5, hampering the amplification of spacer 5 by loss of primer DVRb hybridization site. (e) Simplified spoligotyping membrane results obtained for the five strains and example of a real spoligotyping membrane. Note that the genome spacer order/number does not correspond to the membrane spacer order/number
The polymorphic property of CRISPR arrays, revealed by the presence or absence of certain spacers, has been used for identification and genotyping of clinical isolates of Mycobacterium tuberculosis [7, 8], Streptococcus pyogenes [9], and Campylobacter jejuni [10] contributing to enlighten the epidemiology of the diseases they cause.
A comprehensive and updated database, containing information about CRISPR systems in several bacterial genomes is available on http://crispr.u-psud.fr/crispr/. Convincing CRISPR structures were found in 1,251 genomes out of 2,630 analyzed so far (as of January 2014).
Within the genus Mycobacterium, the presence of CRISPRs structures is exclusive to members of the Mycobacterium tuberculosis complex (MTBC). Their number per genome varies from one (in some strains of Mycobacterium canettii and of M. tuberculosis) to four (in M. tuberculosis H37Rv), with an average of two CRISPRs arrays in all M. bovis analyzed (http://crispr.u-psud.fr/).
In MTBC strains, a CRISPR array consists of multiple 36 bp tandem DRs, with the consensus sequence TGAGGTGCGGC GTGAGCGCGGGT, interspersed by a total of 94 recognized spacers with 35–41 bp in length [11, 12]. The order and sequence of the spacers is highly conserved between strains of the MTB complex. However, the size of the DR region and, thereby, the presence of spacers differ significantly between strains. Differences have been shown to be due to deletions of spacer sequences in the DR region by transposition of the insertion sequence IS6110 [11, 13], which is almost invariably present in the DR cluster of MTBC strains, probably driven by homologous recombination between adjacent or distant DRs [7] or by replication slippage.
Based on the nature of the DNA polymorphism in the DR cluster region, two methods of MTBC strain differentiation were developed: direct variable repeat polymerase chain reaction (DVR-PCR) [7], enabling detection and typing of individual M. tuberculosis strains in a single PCR, and spoligotyping (spacer oligonucleotide typing) [8]. This last method, also based on PCR coupled with reverse line blot hybridization, gained wide diffusion, due to its easy execution and high throughput, to assess global MTBC strain diversity. This technique consists in the amplification of the DR region using two inversely orientated primers, one of them labeled with biotin, complementary to the flanking conserved DR sequence (Figs. 1 and 2). DNA spacer sequences in between adjacent DRs and in between more distantly positioned DRs are amplified [8]. Evaluation of the presence or absence of spacers is done by reverse hybridization of the obtained PCR products with a set of selected 43-spacer oligonucleotides covalently linked to a nylon membrane (Table 1, Fig. 2), designed according to the sequences of M. tuberculosis H27Rv and M. bovis BCG P3 strains. The PCR products are applied onto the membrane in reverse orientation of the rows with the synthetic oligonucleotides. Since one of the DR primers is labeled with biotin, the detection of hybridization is done by chemiluminescence, through addition of streptavidin-peroxidase conjugate and a substrate (Fig. 2).
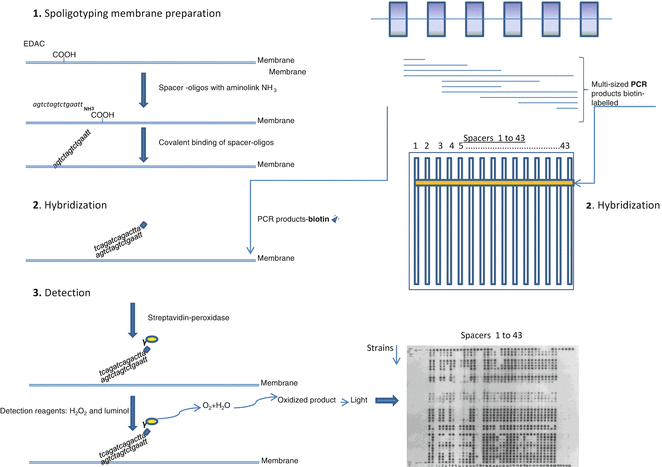

Fig. 2
Schematic basic steps of spoligotyping method. See text for details
Table 1
Sequences and concentrations of the oligonucleotide probes covalently linked to the Biodyne C membrane [8]
Oligonucleotide number order | Sequence (3′-5′ amino) | Concentration (pmol/150 μl) |
|---|---|---|
1 | ATAGAGGGTCGCCGGTTCTGGATCA | 12.5 |
2 | CCTCATAATTGGGCGACAGCTTTTG | 30.0 |
3 | CCGTGCTTCCAGTGATCGCCTTCTA | 12.5 |
4 | ACGTCATACGCCGACCAATCATCAG | 12.5 |
5 | TTTTCTGACCACTTGTGCGGGATTA | 12.5 |
6 | CGTCGTCATTTCCGGCTTCAATTTC | 12.5 |
7 | GAGGAGAGCGAGTACTCGGGGCTGC | 25.0 |
8 | CGTGAAACCGCCCCCAGCCTCGCCG | 50.0 |
9 | ACTCGGAATCCCATGTGCTGACAGC | 12.5 |
10 | TCGACACCCGCTCTAGTTGACTTCC | 15.0 |
11 | GTGAGCAACGGCGGCGGCAACCTGG | 30.0 |
12 | ATATCTGCTGCCCGCCCGGGGAGAT | 60.0 |
13 | GACCATCATTGCCATTCCCTCTCCC | 12.5 |
14 | GGTGTGATGCGGATGGTCGGCTCGG | 30.0 |
15 | CTTGAATAACGCGCAGTGAATTTCG | 30.0 |
16 | CGAGTTCCCGTCAGCGTCGTAAATC | 12.5 |
17 | GCGCCGGCCCGCGCGGATGACTCCG | 100.0 |
18 | CATGGACCCGGGCGAGCTGCAGATG | 12.5 |
19 | TAACTGGCTTGGCGCTGATCCTGGT | 12.5 |
20 | TTGACCTCGCCAGGAGAGAAGATCA | 12.5 |
21 | TCGATGTCGATGTCCCAATCGTCGA | 25.0 |
22 | ACCGCAGACGGCACGATTGAGACAA | 12.5 |
23 | AGCATCGCTGATGCGGTCCAGCTCG | 50.0 |
24 | CCGCCTGCTGGGTGAGACGTGCTCG | 50.0 |
25 | GATCAGCGACCACCGCACCCTGTCA | 25.0 |
26 | CTTCAGCACCACCATCATCCGGCGC | 12.5 |
27 | GGATTCGTGATCTCTTCCCGCGGAT | 25.0 |
28 | TGCCCCGGCGTTTAGCGATCACAAC | 12.5 |
29 | AAATACAGGCTCCACGACACGACCA | 12.5 |
30 | GGTTGCCCCGCGCCCTTTTCCAGCC | 12.5 |
31 | TCAGACAGGTTCGCGTCGATCAAGT | 12.5 |
32 | GACCAAATAGGTATCGGCGTGTTCA | 25.0 |
33 | GACATGACGGCGGTGCCGCACTTGA | 100.0 |
34 | AAGTCACCTCGCCACACCGTCGAA | 25.0 |
35 | TCCGTACGCTCGAAACGCTTCCAAC | 12.5 |
36 | CGAAATCCAGCACCACATCCGCAGC | 12.5 |
37 | CGCGAACTCGTCCACAGTCCCCCTT | 12.5 |
38 | CGTGGATGGCGGATGCGTTGTGCGC | 25.0 |
39 | GACGATGGCCAGTAAATCGGCGTGG | 25.0 |
40 | CGCCATCTGTGCCTCATACAGGTCC | 12.5 |
41 | GGAGCTTTCCGGCTTCTATCAGGTA | 12.5 |
42 | ATGGTGGGACATGGACGAGCGCGAC | 25.0 |
43 | CGCAGAATCGCACCGGGTGCGGGAG | 50.0 |
Spoligotyping applied to bacterial isolates is simple, robust, and highly reproducible. On the other hand, for MTBC strains containing few copies of the IS6110 element, such as M. bovis, spoligotyping is generally more discriminatory than IS6110 typing methods, and the DR region is, consequently, also more stable. A hybridization profile is generated for any MTBC isolate (Fig. 1) allowing, simultaneously, its identification and genotyping, since each species has a specific spoligotype signature [14]. Such is the case of M. bovis, M.canettii, M. caprae, and M. tuberculosis (Table 2). The typical signature of most M. bovis strains and all M. bovis BCG is the absence of spacers 3, 9, 16, and 39–43 [14, 15] (Table 2).
The spoligotyping profile can be expressed as a numerical profile (binary or octal code), which simplifies storing of typing data and facilitates interlaboratory comparison. Most relevant for the global comparison of data is the existence of international spoligotyping databases: SpolDB4 [17] and Mbovis.org [18], the latter specific for M. bovis and other non-M. tuberculosis isolates, characterized by absence in the genome of RD9 (region of difference). Most spoligotype attribution codes use the international databases calls: ST (for shared types) followed by a number in M. tuberculosis databases or SB (for spoligotype bovis) followed by four digits in Mbovis.org database. These databases allow easy interlaboratory comparison of spoligotype patterns and contribute for the definition of the major lineages within M. tuberculosis and M. bovis strains.
In an attempt to enlarge the discriminatory power of spoligotyping, additional spacers have been evaluated for M. bovis [12, 19], M. africanum [16], and M. caprae [20] strains typing. However, these extended spoligotypes have had few practical applications, since other methods, such as mycobacterial interspersed repetitive units variable number of tandem repeats (MIRU-VNTR) in conjunction with spoligotyping, proved to be the most suitable tools to increase the discriminatory power and to further differentiate strains of the MTBC [21, 22].
Provided that suitable quality control measures are adopted, the spoligotyping procedure described below, using 43 spacer analyses, should enable the identification and typing of members of the MTBC. For a self-sufficient procedure and independent from commercial membranes, we describe the in-house preparation of the spoligotyping membrane.
2 Materials
All the solutions should be freshly prepared, with ultrapure water (18.2 MΩ cm at 25 °C) and analytical grade reagents, and stored at room temperature. The stock solutions should be sterilized in autoclave at 121 °C for 15 min. The nylon membrane can be regenerated and reused up to ten times.
2.1 Solutions
2.1.1 Stock Solutions
1.
0.5 M EDTA: For 1,000 ml, weight 186.12 g of ethylenediaminetetraacetic acid (EDTA) and add half the water. Mix until complete dissolution using a magnetic stirrer and adjust to pH 8.0. Complete the volume to 1,000 ml (see Note 1 ).
2.
20× SSPE (w/v): Weight 35.6 g of Na2HPO4.2H2O, 210.24 g of NaCl, and 7.4 g of EDTA for a final volume of 1,000 ml in water. Mix well and adjust to pH 7.4.
2.1.2 Working Solutions
For Membrane Preparation
1.
500 mM NaHCO3 (pH 8.4): For 250 ml, weight 10.5 g of NaHCO3 and dissolve in water. Mix, adjust to pH 8.4, and add water up to 250 ml.
2.
16 % (w/v) EDAC: For 10 ml, add 1.6 g of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDAC) and mix with 9.4 ml of water. Store at −20 °C.
3.
100 mM NaOH: Weight 1.0 g of sodium hydroxide (NaOH) and add water up to 250 ml and mix.
4.
10 % SDS (w/v): Weight 10 g of sodium dodecyl sulfate (SDS) to a glass bottle, and add water up to a final volume of 100 ml (see Note 2 ).
5.
2× SSPE/0.1 % SDS (w/v): For 250 ml solution, add 25 ml of 20× SSPE to 222.5 ml of water. After stirring, carefully add 2.5 ml of 10 % SDS (w/v) (see Note 3 ).
6.
1:100 China ink diluted in 2× SSPE: For 300 μl, add 3 μl of China ink to 297 μl of 2× SSPE.
7.
20 mM EDTA: For 100 ml, add 4 ml of 0.5 M EDTA to 96 ml of water.
For Hybridization
1.
10 % (w/v) SDS: For 130 ml, add 13 g of SDS to 100 ml of water. Stir until complete dissolution using a magnetic stirrer. In a graduated cylinder, add water up to 130 ml final volume (see Note 2 ).
2.
2× SSPE (v/v): For 500 ml, add 50 ml of 20× SSPE to 450 ml of water.
3.
2× SSPE/0.1 % SDS: For 300 ml, add 30 ml of 20× SSPE to 267 ml of water. After stirring, carefully add 3 ml of 10 % (w/v) SDS (see Note 3 ).
4.
2× SSPE/0.5 % SDS: For 1,000 ml, add 100 ml of 20× SSPE to 850 ml of water. After stirring, carefully add 50 ml of 10 % SDS (see Note 3 ).
5.
1 % SDS: For 750 ml, add 75 ml of 10 % SDS to 675 ml of water.
6.
20 mM EDTA: For 1,000 ml, measure 40 ml of 0.5 M EDTA and add water up to 1,000 ml.
2.2 Membrane Preparation
1.




Oligonucleotide probes synthesized with a 5′-end amino group (see Note 4 ), diluted in 150 μl of 500 mM NaHCO3 pH 8.4.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree



